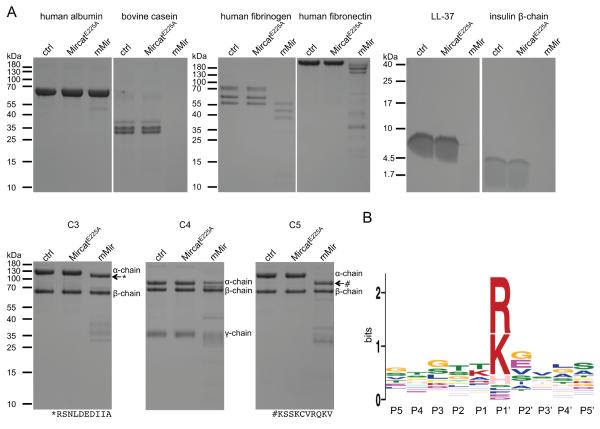
Figure 5.
Mirolysin hydrolyzes a broad range of substrates and cleaves peptide bonds at arginine and lysine. A) The proteins human albumin, fibrinogen, fibronectin, and bovine casein, the human complement proteins C3, C4, and C5 (10 μg), and the peptides human cathelicidin LL-37 and bovine insulin β-chain (4 μg) were incubated in 50 mM Tris, 150 mM NaCl, 2.5 mM CaCl2, 0.02% NaN3, pH 7.5, at 37°C for 8 h alone (control, ctrl) or with mMir at a substrate:enzyme weight ratio of 1:20 (complement proteins), 1:100 (other proteinaceous substrates), and 1:1 000 (peptides). Substrates incubated with MircatE225A served as a negative control. The identity of the bands indicated by arrows was determined by N-terminal sequencing. The obtained sequences are shown below the gels. B) The digestive peptides resulting from cleavage of substrates by mirolysin were identified by mass spectrometry, thus allowing the determination of cleavage sites within the substrate polypeptide chains. Based on these results, the mirolysin cleavage motif was generated employing MEME (Multiple Em for Motif Elicitation).