Figure 4.
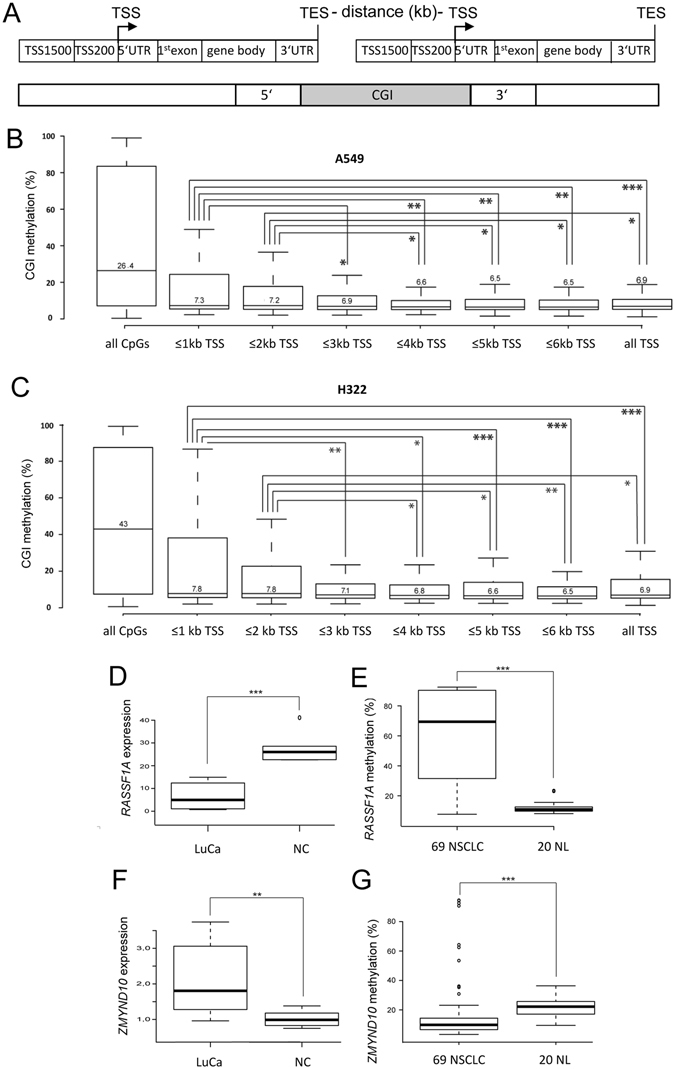
Epigenetic silencing of downstream genes located in tandem orientation. (A) Outline of the organization of tandem oriented genes with a CpG island at the transcriptional end site (TES) of the upstream gene and the transcriptional start site (TSS) of the downstream gene. (B) Methylation level of TSS-associated CGI in A549 cells was plotted depending on the distance between TES and TSS in kb (n = 258 to 297 CGI). Methylation as detected using 450 K bisulfite bead chip arrays. “All CpGs” and “all TSS” indicate the methylation level of CpGs located at CGI and TSS-associated CGI, respectively. (C) Methylation levels of tandem oriented downstream CGI in H322 cells. (D) RASSF1A expression levels were analyzed in lung cancer cell lines (LuCa: A427, A459, H322 and H358) and normal lung cells (NC: PAF and PASMC) by qRT-PCR. RASSF1A expression was normalized to the GAPDH level. (E) Methylation of RASSF1A was analyzed in 69 non small cell lung cancer (NSCLC) cell lines and 20 normal lung tissues (NL). For details see also Fig. 3. (F) ZMYND10 expression levels were analyzed in LuCa and NC as measured by qRT-PCR and normalized to GAPDH levels. (G) Methylation of ZMYND10 was analyzed in 69 NSCLC cell lines and 20 NL. *p < 0.05, **p < 0.01 and ***p < 0.001.
