Figure 6.
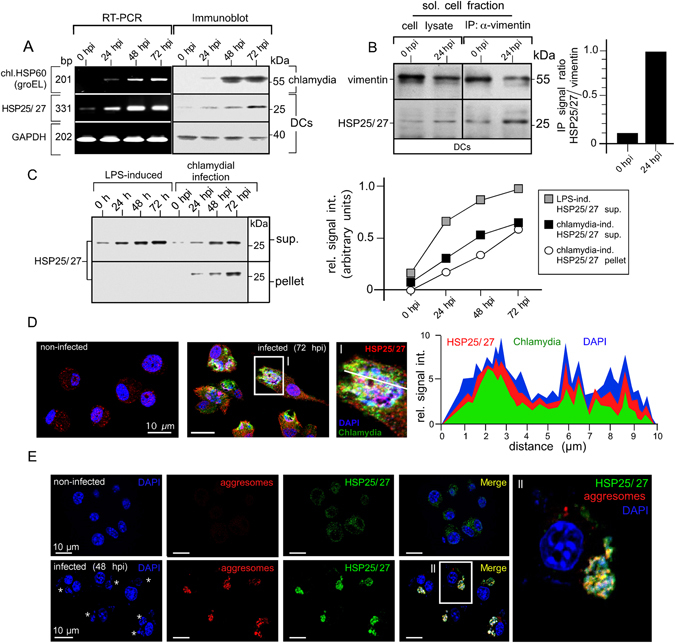
Chlamydial structures interact with HSP25/27 involved in aggresome formation and clearance. (A) DCs were infected and total RNA was analysed by semi-quantitative RT-PCR for HSP25/27, chl.HSP60 and GAPDH (left panel). Corresponding cell lysates were analysed in Western blot (right panel). (B) DCs were infected or not with chlamydia for 24 h. The soluble fraction of lysates was subjected to immunoprecipitation of vimentin. Western blots were probed for vimentin and HSP25/27 (left panel). Signal quantification was performed by densitometry (right panel). (C) DCs were treated with LPS or infected with chlamydia. Cells were disrupted using a Dounce homogeniser and centrifuged. Pellet and supernatant fraction were analysed by Western blot (left panel). HSP25/27 signals were determined by densitometric scanning and plotted in a respective graph as arbitrary units (right panel). (D) Non-infected and infected (72 hpi) DCs were costained with antibodies specific for HSP25/27 (red), chlamydial LPS (green) and DNA (blue) (left panel). The enlarged photograph (I) shows colocalisation between HSP25/27 and chlamydial structures. The fluorescence intensity along a cellular cross section of interest was measured (right panel). Obtained profiles were overlaid and coloured. (E) Infected and non-infected DCs (48 hpi) were costained for HSP25/27 (green), aggresomal structures (red) and DNA (blue). The enlarged photograph (II) shows colocalisation between HSP25/27 and aggresomal structures.
