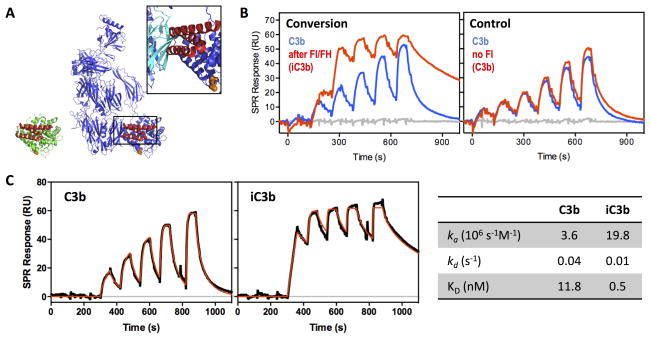
Figure 6. Differential accessibility of Efb-C binding sites in C3b and iC3b.
A. Crystal structure of C3d:Efb-C (N138A mutant; PDB 3D5R; (27)) with C3d in green and Efb-C in red on the left and a model in which the TED domain of C3b (blue; PDB 2I07; (4)) is aligned with C3d. The insert shows the steric clash of Efb-C with the MG1 domain of C3b (highlighted in cyan). B. In agreement with the hypothesis of a disordered and more flexible CUB domain in iC3b, Efb-C binds more strongly to surface-immobilized iC3b than to C3b, as determined by SPR. In the semi-quantitative experiment, thioester-biotinylated C3b was captured on a streptavidin-coated sensor chip and a titration was performed with Efb-C (3–100 nM) before and after conversion of one C3b surface to iC3b to demonstrate that the observed increase in activity was indeed due to the iC3b transformation with loss of steric restriction around the Efb-C binding site. C. In order to quantitate the differential interaction profile, a kinetic titration was performed on the C3b and iC3b surfaces using consecutive injections of Efb-C at increasing concentration (6–100 nM), and the resulting SPR curves (black) were fitted to a Langmuir 1:1 binding model (red lines) to determine kinetic rate constants and binding affinity (table on the right). iC3b showed a >20-fold higher affinity for Efb-C when compared to C3b, caused by a ~5-fold higher association rate and a ~4-fold slower dissociation rate. Of note, a single amino acid mutant of Efb-C (i.e., N138A) that shares the same binding mode as wildtype Efb-C but features lower complex stability (KD of 19.8 vs. 1.5 nM; (27)) was used to facilitate the experiment and avoid harsh regeneration conditions.