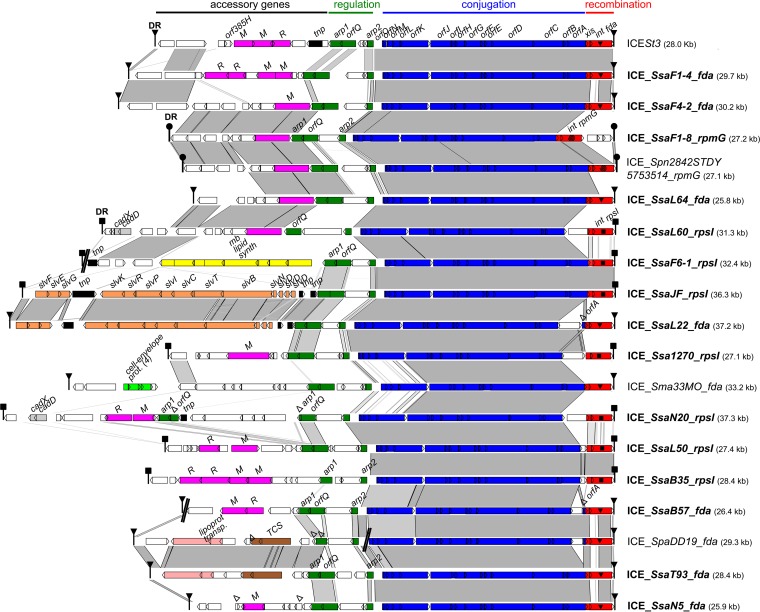
FIG 1.
Comparison of the integrative and conjugative elements (ICEs) found in S. salivarius and in other streptococci. ICEs are named according to their host strains and integration sites. ICEs of S. salivarius are indicated in bold. For more clarity, elements in accretion with ICEs are not shown. Nucleic acid sequence identity higher than 80% between sequences is indicated in light gray and that higher than 90% in dark gray. Direct repeats (DR) delimiting ICEs are shown as triangles, circles, or squares depending on their sequences. Open reading frames (ORFs) appear as arrows (truncated genes are indicated by capital deltas). Modules of recombination (integrase [int] and excisionase [xis] genes), conjugation (orfO to orfA genes), and regulation (including the arp1, arp2, and orfQ genes) appear in red, blue, and green, respectively. The three different integration genes (fda, rpsI, or rpmG) targeted by the integrase are indicated by distinct symbols in the integrase gene and are part of the ICE name. Genes from the adaptation module encoding proteins with putative function inferred from in silico analysis are indicated in pink for RM systems and orphan methyltransferase genes, in dark gray for cadmium resistance genes (cadD and cadX), in yellow for the membrane lipid synthesis cluster, in orange for the bacteriocin synthesis cluster, in light green for genes encoding cell envelope proteins, in light pink for the cluster of genes for putative lipoprotein transport, in brown for the two-component system (TCS), and in black for the tnp transposase gene(s). Sequences used for this analysis were ICESt3 of S. thermophilus (AJ586568) and sequenced genomes of S. pneumoniae 2842STDY5753514 ( FDNK01000013), S. macedonicus 33MO (JNCV01000015), and S. parasanguinis DD19 (LQNY01000339, LQNY01000340). Gaps in the assembly found in ICE_SsaF6-1_rpsI, ICE_SsaB57_fda, and ICE_SpaDD19_fda are indicated by a double slash.