Fig. 1.
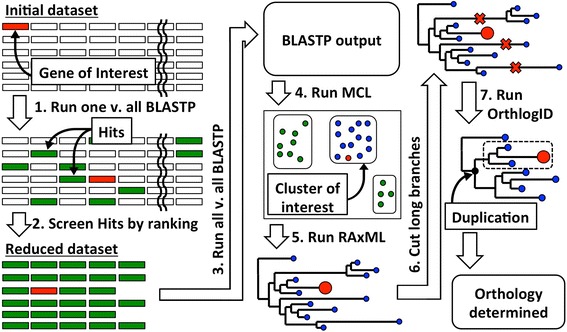
OrthoReD overview. To determine the orthology of the gene of interest, gene of interest is used as a query for a BLASTP search against the dataset (step 1). The BLASTP hits are screened to generate a reduced dataset (step 2). All-v-all BLASTP is conducted on the reduced dataset (step 3) to generate pairwise similarity matrix used by MCL to separate the reduced dataset into clusters (step 4). Most likely phylogeny is reconstructed for the members within the cluster of interest (step 5) and long branches are subsequently removed from the tree (step 6). Finally, all members of the clade that share the most recent gene duplication event are returned as predicted orthologs (step 7)
