Figure 1.
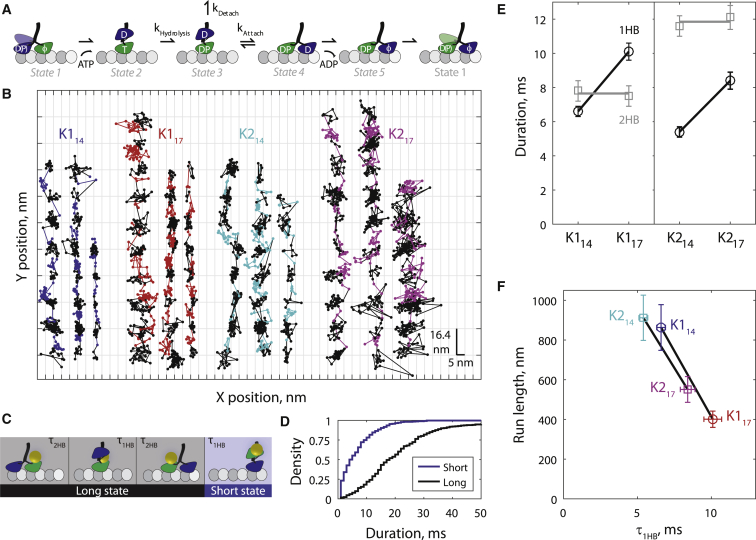
Extending the NL reduces processivity by slowing the tethered head attachment rate. (A) Five-state mechanochemical cycle established in Mickolajczyk et al. (28) and Chen et al. (34), in which processivity is set by a kinetic race between attachment of the tethered (blue) head and detachment of the bound (green) head in the 1HB posthydrolysis state (state 3). Here T denotes ATP, D denotes ADP, DP denotes ADP plus phosphate, and φ denotes no nucleotide. (B) Example 1000 frames/s traces of kinesin-1 (K1) and kinesin-2 (K2) motors with 14- and 17-aa NL domains stepping in 2 mM ATP. Horizontal lines denote inferred microtubule binding sites of the nanoparticle-labeled head. Long states on the microtubule (black points) and short states off the microtubule (colored points corresponding to motor type) were determined by fitting to the (X,Y,t) data (details in Supporting Material). Each data point represents 1 ms. (C) Two-state mechanical model in which each 16.4-nm step is separated into 1HB and 2HB states. Long and short states correspond to black and colored points, respectively, in (B). Note: the terms “long” and “short” refer to duration, not molecular configuration. (D) Distributions of N = 370 long states and N = 347 short states for K114; see Fig. S2 for other motor distributions. (E) 1HB and 2HB durations showing that for both kinesin-1 and kinesin-2, lengthening the neck linker from 14 to 17 aa increased the 1HB duration and did not change the 2HB duration. Values were calculated from means of long and short state distributions. Error bars represent propagated error from the SE of long and short state distributions. (F) Run length is negatively correlated with 1HB duration for both kinesin-1 and -2, indicating that motors with long NL have reduced processivity due to a reduced kAttach. Colors denoting motor type correspond to (B). To see this figure in color, go online.