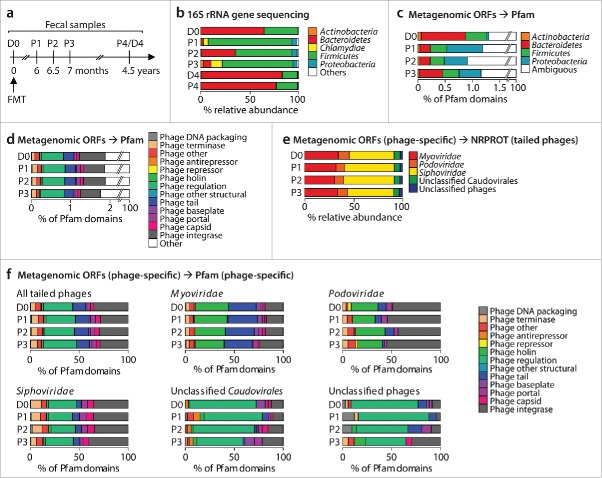
Figure 1.
(a) Timeline of fecal samples of donor (D0 and D4) and patient (P1 through P4) subjected to 16S rRNA gene sequencing (all samples) or metagenomic sequencing (samples D0 and P1-P3). (b) Relative abundances of bacterial phyla as inferred by 16S rRNA gene sequencing. (c) and (d) Relative proportions of bacterial phylum-specific (c) or phage-specific (d) Pfam domains identified from metagenomic ORFs. (e) Relative abundances of phage families identified by aligning phage-specific ORFs against tailed phage protein sequences of the NRPROT database. (f) Virus family-specific relative proportions of phage-specific Pfam domains identified from metagenomic ORFs.