Figure 1.
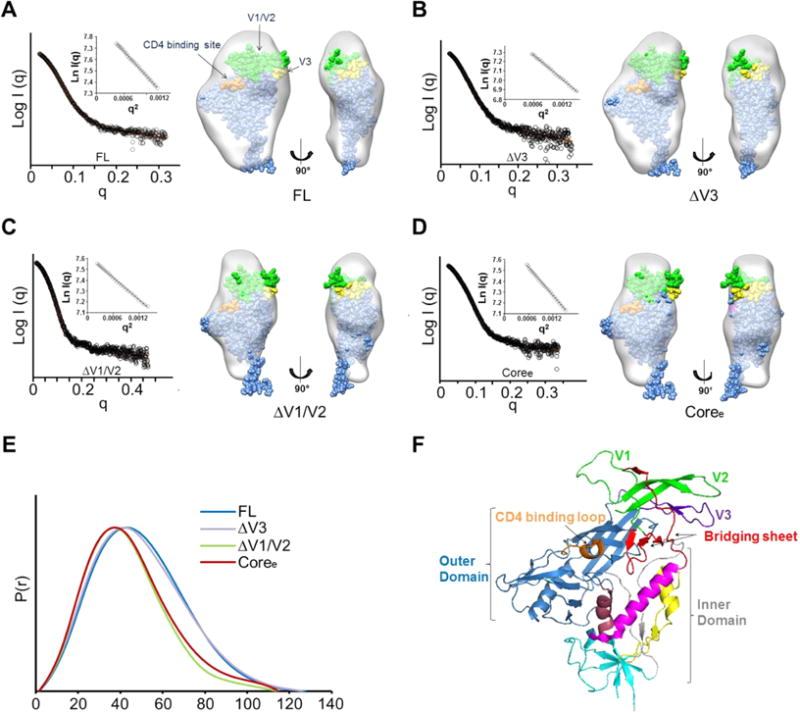
Global gp120 subunit structures determined by small angle X-ray scattering. (A) SAXS pattern (black) for unliganded YU2 FL. The orange line represents the SAXS pattern from a DAMMIN ab initio SAXS reconstruction. Insets show the linear Guinier fit. DAMMIN reconstruction for FL. The structure of the gp120 monomer from the BG505 SOSIP trimer structure (PDB entry 4TVP)93 was fitted into the SAXS density. The V1/V2 residues are colored green. The V3 residues are colored yellow. The contact residues at the CD4-binding site are colored orange. (B) SAXS pattern (black) for unliganded ΔV3 and DAMMIN model for ΔV3. (C) SAXS pattern (black) for unliganded ΔV1/V2 and DAMMIN model for ΔV1/V2. (D) SAXS pattern (black) for unliganded Coree and DAMMIN model for Coree. ΔV3, ΔV1/V2, and Coree SAXS models show a loss of crown density leading to a poorer fit of the V1/V2 and V3 regions in the gp120 subunit as organized in the SOSIP trimer crystal structure. (E) Pairwise distance distributions [P(r)] for FL (blue), ΔV3 (purple), ΔV1/V2 (green), and Coree (red) show changes in size consistent with variable loop truncation. (F) HIV gp120 architecture (PDB entry 4NCO). Different regions in gp120 are highlighted by color: outer domain (blue); inner domain (gray), including layer 1 (yellow), layer 2 (pink), layer 3 (dark red), and seven-stranded β-sandwich (cyan); bridging sheets (red); CD4-binding loop (orange); V1/V2 loop (green); and V3 loop (purple).
