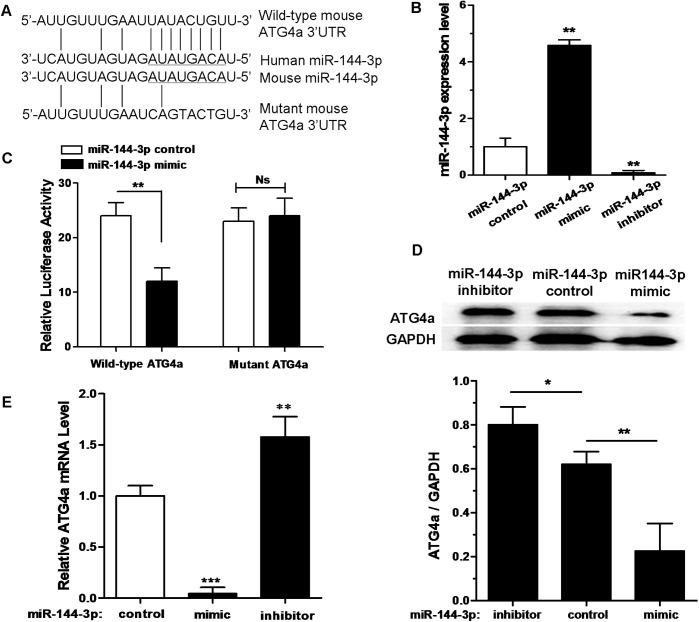
Fig 2. miR-144-3p represses ATG4a expression by targeting its 3' UTR.
(A) Bioinformatic analysis was used to predict the mouse miR-144-3p seed sequences and the 3'UTR sequences of wild type (WT) and mutant ATG4a. (B) The expression level of miR-144-3p in RAW264.7 cells transfected with miR-144-3p control, miR-144-3p mimic or its inhibitor was determined by a qRT-PCR assay. qRT-PCR data were normalized with U6 small nucleolus RNA data. Data represent the mean ± SD from three independent experiments. **p<0.01. (C) RAW264.7 cells were cotransfected with pMIR-REPORT Luciferase vector carrying wild-type ATG4a or mutant ATG4a, along with miR-144-3p control or miR-144-3p mimics, and a luciferase assay was performed. Data are shown as mean ± SD of four independent experiments. **p<0.01. (D) RAW264.7 cells were transfected with miR-144-3p control, miR-144-3p mimic or miR-144-3p inhibitor. 50 μg of total cell extracts were analyzed by Western blotting with a rabbit anti-ATG4a antibody. Representative Western blot image for ATG4a expression and quantitative analysis of ATG4a protein levels, normalized to GAPDH expression, are shown. Data are shown as mean ± SD of four independent experiments. *, p <0.05; **, p<0.01. (E) RAW264.7 cells were transfected with miR-144-3p control, miR-144-3p mimic or miR-144-3p inhibitor. The expression levels of ATG4a mRNA were detected by qRT-PCR. Data represent the mean ± SD from three independent experiments. **p<0.01, ***p<0.001.