Fig. 5. Condensin II and TFIIIC are colocalized at TAD boundaries.
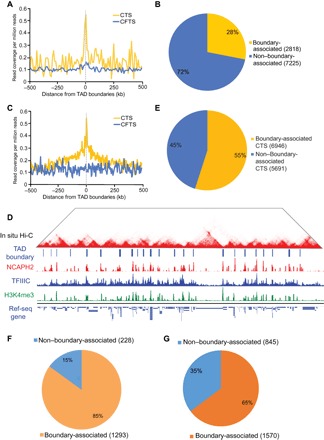
(A) CTS, but not CFTS, were enriched at TAD boundaries in mESCs. (B) Twenty-eight percent of CTS were within 50 kb of a TAD boundary in mESCs, in contrast to 13% TAD boundary association of random control peaks by chance (P = 2.1 × 10−16, χ2 test). (C) CTS, but not CFTS, were enriched at TAD boundaries in human HEK293 cells. (D) Representative image from a genome browser showing tracks of in situ Hi-C and ChIP-seq of NCAPH2, TFIIIC-220, and H3K4me3. (E) Fifty-five percent of CTS were within 50 kb of a TAD boundary in mESCs, in contrast to 41% TAD boundary association of random control peaks by chance (P = 2.11 ×10−8, χ2 test). (F) The total number of genes that are significantly down-regulated upon condensin knockdown is 1516 in mESCs. Eighty-five percent (1293) of genes that were significantly down-regulated upon NCAPH2 knockdown were within 50 kb of a TAD boundary. (G) A total of 2415 genes are significantly down-regulated upon condensin knockdown in HEK293 cells. Sixty-five percent (1570) of genes that were significantly down-regulated upon NCAPH2 knockdown were within 50 kb of a TAD boundary.
