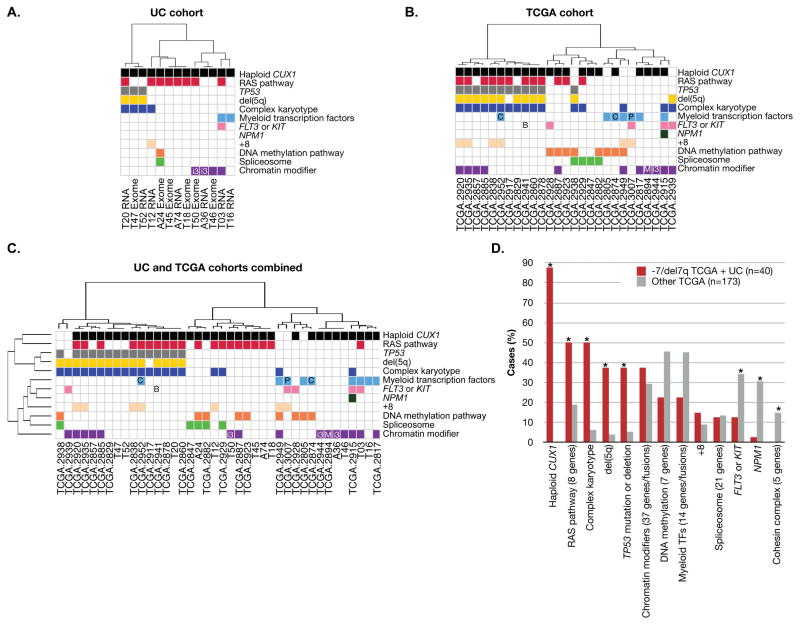
Figure 1.
The pattern of somatic mutations in -7/del(7q) leukemias is distinct from other AML types. Categorization of genes within pathways is as defined (TCGA 2013) (Table S4). Mutations in genes not in these pathways are not shown. Samples are hierarchically clustered by Pearson correlation coefficients based on the presence or absence of mutations in these pathways using Ward’s method. Mutated pathways are shown for the UC cohort (A), the TCGA cohort (B), and the combined UC and TCGA cohorts (C). D. The frequency of the alteration in the combined UC (n=13) and TCGA (n=27) cohorts of ‒7/del(7q) leukemias (red bars, n=40) is shown in comparison to TCGA AML samples without -7/del(7q) (grey bars, n=173). The number of genes per category is indicated in parentheses. * indicates chi-squared test p < 0.05 comparing -7/del(7q) TCGA samples versus other TCGA samples. All recurrent genetic abnormalities according to the 2008 WHO classification “AML with recurrent genetic abnormalities” are indicated (Swerdlow et al. 2008), with an abbreviation within the relevant pathway. B: BCR-ABL fusion; C: CEBPA mutation; i3: inv(3)(q21q26.2) or t(3)(3;3)(q21;q26.2); M: MLLT3-MLL fusion; and P: PML-RAR fusion. Abbreviations: TF, transcription factor. Within the UC cohort, t-MN samples are named by TXX and de novo AML samples are named by AXX.