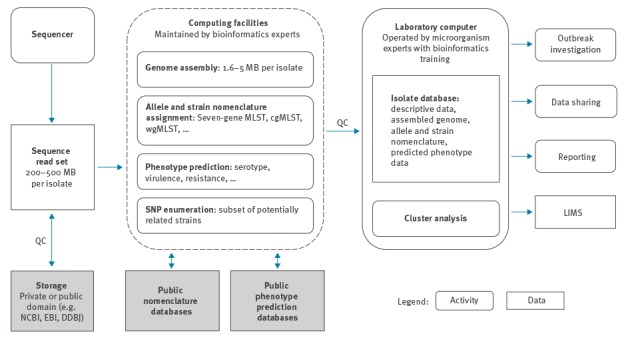
Figure 2.
Potential solutions for computing and storage in PulseNet International laboratories, May 2017
Cg: core genome; DDBJ: DNA Data Bank of Japan; EBI: European Bioinformatics Institute; LIMS: Laboratory Information Management System; MB: megabyte; MLST: multilocus sequence typing; NCBI: National Center for Biotechnology Information; QC: quality control; SNP: single nucleotide polymorphism; wg: whole genome.
This workflow separates storage of the sequence data, from the intensive bioinformatics analyses that need to be performed on them, in particular genome assembly and allele calling. This allows performing these analyses on a dedicated computing cluster either in-house or in the cloud, where they can be maintained and updated by bioinformatics experts who may not reside within the same organisation or even country as the PulseNet laboratory. No confidential information is handled on this computing cluster. Within the PulseNet laboratory, microorganism experts will perform quality controls, cluster analysis on MLST data, as well as evaluating virulence and other factors. Following data interpretation, laboratory experts will use this information for outbreak investigations, reporting and sharing with other agencies. When required, SNP analysis will be performed on subsets of closely related strains.