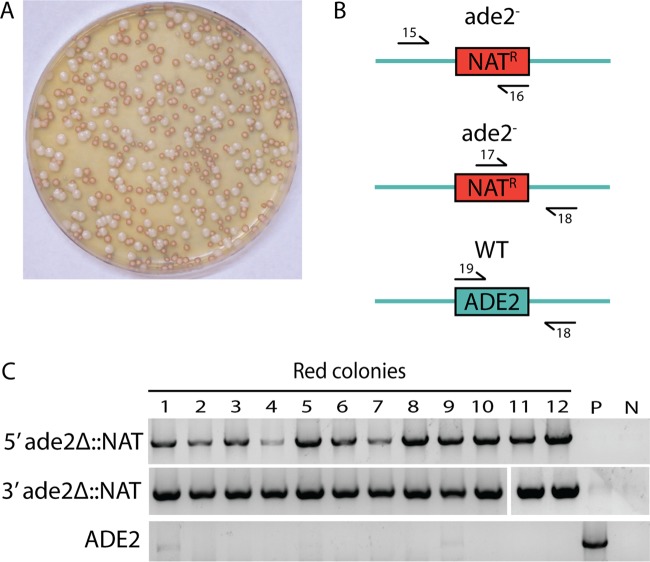
FIG 2 .
Genotypic analysis of red colonies arising from CRISPR-Cas9 targeting of ADE2 in a C. lusitaniae haploid strain. (A) A C. lusitaniae-optimized CRISPR-Cas9 system was used to target the ADE2 locus, and a large percentage of red-colony (ade−) phenotypes was observed (gamma of image adjusted to emphasize red/white color differences). (B) PCR assays for identifying ade2 genotypes, with arrows indicating the relative positions of the primers used (see Data Set S1 for sequences). WT, wild type. (C) For all 12 red colonies shown here, the 5′ and 3′ junction checks were positive and the ORF checks were negative, indicating successful replacement of the ADE2 target gene with the nourseothricin resistance marker. The parent strain (RYS284) was used as a wild-type control (P), and template DNA was omitted in the negative control (N).