Fig. 1.
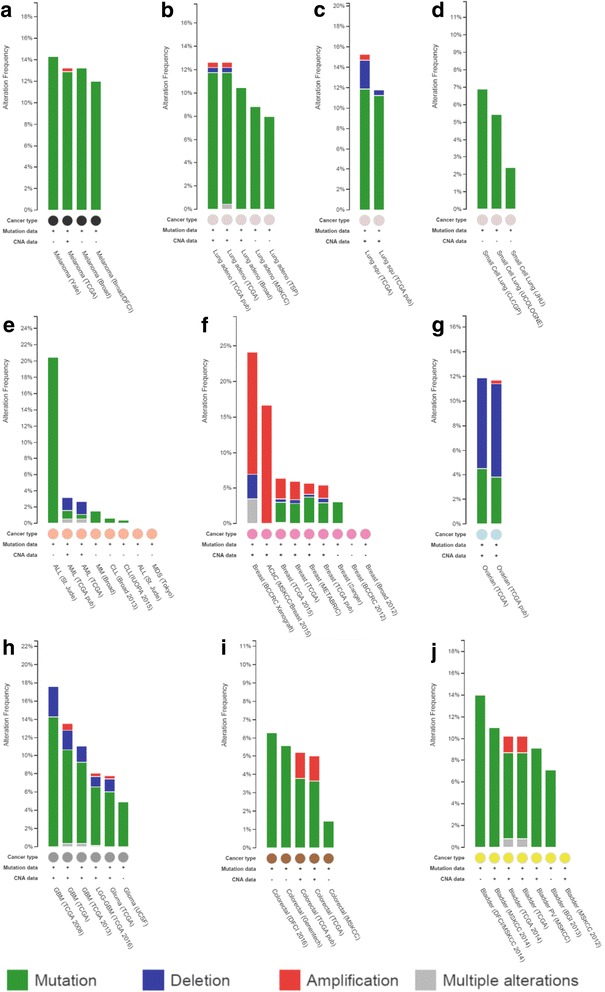
The frequency and nature of somatic NF1 mutations in different cancer types derived from the cBio dataset. a Malignant melanoma. b Lung adenocarcinoma. c Lung squamous cell carcinoma. d Small cell lung carcinoma. e Acute lymphocytic leukaemia (ALL). Acute myeloid leukaemia (AML), chronic lymphocytic leukaemia (CLL), malignant myeloma (MM) and myelodysplastic syndrome (MDS). f Breast carcinoma. g Serous ovarian carcinoma. h Brain glioma, including glioblastoma multiforme (GBM). i Colorectal carcinoma. j Bladder transitional cell carcinoma. Mutations = single base-pair substitutions, in-frame microdeletions/insertions, frameshift microdeletions/insertions, splice site mutations (including those that can create in-frame deletions via exon skipping), nonsense mutations and frameshift insertions/deletions (shown in green); deletions = gross, multi-exonic and whole gene deletion identified as copy number changes (shown in blue); amplification = multi-exonic, whole gene duplications identified as copy number changes (shown in red); multiple alterations = some combination of mutations, deletions and/or amplification (shown in grey) [44–46]
