FIG. 1.
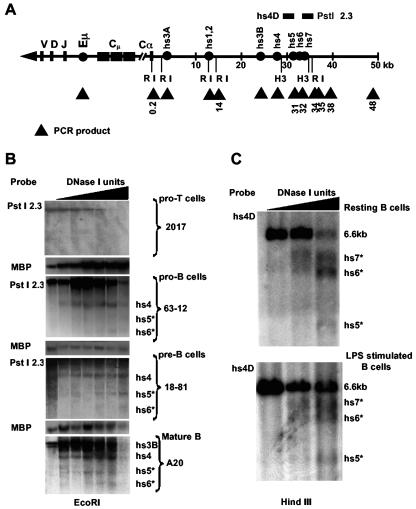
DNase I hypersensitive sites in the region downstream of hs4. (A) Schematic map of the murine Igh locus shows V, D, J, and constant region genes (solid bars) and enhancers and DNase I-hypersensitive sites (solid circles). The murine Igh locus is located on chromosome 12 and comprises hundreds of variable (V) genes, 16 diversity (D) genes, and 4 joining (J) genes spanning more than 3 Mb. Between Cμ and Cα, there are additional constant region genes: δ, γ3, γ1, γ2b, γ2a, and ɛ. The numbers represent the position, in kilobases, along the insert in BAC199M11, which begins downstream of the last exon of the Cα gene. EcoRI (RI) restriction enzyme sites are depicted, as are HindIII (H3) sites used for analysis of DNase I hypersensitivity. The horizontal bars above the line in the vicinity of hs4 represent the PstI-2.3 and hs4D probes used in this study. The PstI-2.3 probe detects a ∼22-kb RI fragment (BALB/c), while hs4D detects a ∼6.6-kb HindIII fragment (C57BL/6). Triangles are targets used for ChIP analysis. (B) EcoRI digestion of DNase I-treated nuclei from the 63-12 pro-B cell line, the 18-81 pre-B cell line, the A20 mature B cell line, and the 2017 pro-T cell line. The ∼22-kb fragment detected by PstI-2.3 includes hs4 as well as two novel DNase I-hypersensitive sites, hs5 and hs6 (*), located ∼4.5 and 3.5 kb, respectively, upstream of the distal EcoRI site. (C) HindIII digestion of DNase I-treated nuclei from resting and LPS-stimulated B cells revealed an additional hs4 downstream DNase I-hypersensitive site, hs7 (*).