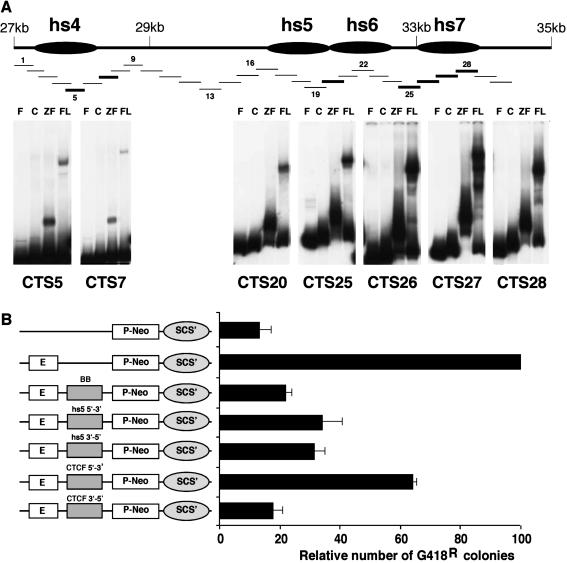
FIG. 6.
Identification and analysis of CTCF sites in the region downstream of hs4. (A) In-scale schematic of the 3′ RR showing 30 consecutive overlapping DNA fragments (fragment numbers intermittently indicated) used for EMSA. EMSA identified CTCF binding sites associated with hs4, hs5, and the region downstream of hs6, i.e., hs7. F, free DNA probe; C, control TnT reaction with luciferase template; ZF, 11 zinc finger region of CTCF; FL, full-length CTCF. (B) Assessment of insulator activity of hs5 and the region containing strong CTCF binding sites (hs7) (see Materials and Methods for exact fragments tested) by using a stable transfection assay that measures NeoR colonies. The number of colonies detected with the construct containing both the T-cell receptor enhancer and the promoter ranged from ∼130 to 180 in different experiments. We set this number at 100 and normalized all the other data accordingly. BB identifies the BEAD-1 insulator and is used as a positive control.