Abstract
The mutated in colorectal cancer (MCC) gene is an important colorectal tumor suppressor gene, although few studies have reported the microRNA(s) that could directly target MCC in colorectal cancer. Here, we used microRNA (miRNA) target prediction algorithms, and previously reported microarray data in human colorectal cancer found that only miR-4261 was predicted by all three databases to directly target MCC. Based on specimens from our own cohort of colorectal cancer patients, we further demonstrated that miR-4261 was overexpressed in colorectal cancer. Interestingly, overexpression of miR-4261 could enhance cell proliferation and G1/S phase transition of cell cycle, and promote cell migration in HCT116 and HT29 cells, while inhibition of miR-4261 had opposite effects. Luciferase reporter assay and western blot analysis confirmed MCC as a direct target of miR-4261. MCC small interfering RNA (siRNA) could abolish the suppressive effects of miR-4261 inhibitor on cell proliferation and migration in HCT116 and HT29 cell lines. Finally, we showed that therapeutic intervention with lentivirus-based miR-4261 sponge injection could effectively reduce tumor growth and inhibit cell proliferation in colorectal cancer xenograft. Collectively, our study is the first one to unravel the functional role of miR-4261, and it provides strong evidence that inhibition of miR-4261 through targeting of MCC might exert a therapeutic effect for colorectal cancer.
Keywords: colorectal cancer, miR-4261, MCC
Introduction
Colorectal cancer is one of the most common malignancies and a leading cause of cancer-related death worldwide.1 It is also a cancer type closely related to genetic and epigenetic alterations.2, 3 Despite the great advances achieved in molecularly targeted agents and chemotherapy for colorectal cancer, many patients do not clinically benefit from treatment because of different tumor mutation types, chemotherapy resistance, and adverse reactions.4, 5 Indeed, the development of novel therapeutic targets for colorectal cancer is still urgently needed.
A class of small non-coding RNAs, termed microRNAs (miRNAs, miRs), have increasingly been reported to be associated with colorectal cancer carcinogenesis.6 Dysregulated miRNAs such as miR-17-5p, miR-320a, miR-143, and miR-21 contribute to colorectal cancer cell proliferation and migration via regulating Wnt/β-catenin, mitogen-activated protein (MAP) kinase, and phosphatidylinositol 3-kinase (PI3K)/Akt pathways.7, 8, 9, 10 Other miRNAs including miR-365, miR-195, and miR-34a are implicated in the control of colorectal cancer cell apoptosis via targeting Bcl-2 and SIRT1, thus activating caspase-3 and caspase-9.11, 12, 13 Moreover, some miRNAs like miR-221, miR-145, miR-103, miR-107, and miR-200c are involved in colorectal cancer metastasis by regulating cell invasion, angiogenesis, and mesenchymal-epithelial transition.14, 15, 16, 17, 18 These miRNAs have been identified to regulate their specific target genes, thus contributing to colorectal cancer carcinogenesis.6
Based on our knowledge about the miRNA-mRNA interactions in colorectal cancer, little is known about the miRNA(s) that could target the mutated in colorectal cancer (MCC) gene, a crucial tumor suppressor of colorectal cancer.19 MCC was originally discovered through its linkage to the adenomatous polyposis coli (APC) gene on chromosome 5q21, and was later identified as a putative colorectal tumor suppressor.20 Compelling evidence indicates that MCC could be mutated and/or dysregulated in human and experimental colorectal cancer.19, 21, 22, 23 MCC was previously reported to block the cell cycle progression from G0/G1 to S phase in mouse NIH 3T3 cells.24 More recently, it was revealed that MCC could inhibit colorectal cancer cell proliferation via inhibiting Wnt/β-catenin signal transduction.25, 26 All of these data imply the significant biological functions of MCC; however, currently, few studies have reported the potential miRNAs that could target MCC in colorectal cancer.
In the present study, we first used miRNA target prediction algorithms (miRDB, miRWalk, and Diana Tools) and previously reported microarray data in human colorectal cancer (accession number GEO: GSE72199),27 and found that among the dysregulated miRNAs detected in this microarray, only miR-4261 was predicted by all the three databases to directly target MCC. miR-4261 was previously reported to be expressed by human embryonic stem cells and neural precursors.28 However, to the best of our knowledge, currently, little is known about the functional roles of miR-4261. Here, we demonstrated that miR-4261 was overexpressed in colorectal cancer tissues from patients clinically diagnosed at the Department of General Surgery of Tongji Hospital. Based on HCT116 and HT29 cell lines, we found that overexpression of miR-4261 could enhance cell proliferation and G1/S phase transition of cell cycle, and promote cell migration of colorectal cancer cells. Importantly, MCC was identified as a direct functional target gene of miR-4261 in colorectal cancer. Finally, we provided in vivo evidence that suppression of miR-4261 had a therapeutic effect on colorectal cancer development. Collectively, inhibition of miR-4261 through targeting of MCC might be a potential therapeutic strategy for colorectal cancer patients.
Results
miR-4261 Is Overexpressed in Human Colorectal Cancer
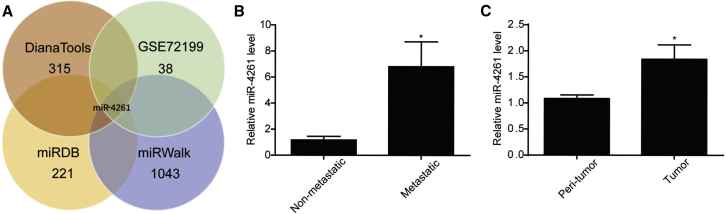
It was recently reported that 38 miRNAs, including miR-4261, were dysregulated in colorectal cancer patients according to the microarray data (GEO accession number: GSE72199).27 In order to identify the potential upstream regulator for MCC, an important tumor suppressor of colorectal cancer, we applied miRNA target prediction algorithms by using miRDB, miRWalk, and Diana Tools databases. Interestingly, among the 38 miRNAs detected by microarray that were dysregulated in colorectal cancer,27 only miR-4261 was predicted by all three databases to directly target MCC (Figure 1A). Thus, miR-4261 was selected to be further examined in our human colorectal cancer specimens. First, we determined miR-4261 expression level based on seven metastatic versus seven non-metastatic human colorectal cancer tissues, which demonstrated a significant upregulation of miR-4261 in metastatic colorectal cancer tissues compared with non-metastatic ones (Figure 1B). Besides that, a total of 42 pairs of cancer tissues versus adjacent normal tissues were collected from patients clinically diagnosed as colorectal cancer at the Department of General Surgery of Tongji Hospital. Using quantitative real-time PCR, we further validated that miR-4261 was overexpressed in colorectal cancer tissues compared with adjacent normal tissues (Figure 1C). Thus, the functional roles of miR-4261 in colorectal cancer were investigated in further detail.
Figure 1.
miR-4261 Is Elevated in Human Colorectal Cancer
(A) miR-4261 is predicted to directly target MCC by miRNA target prediction algorithms. (B) Increased expression level of miR-4261 in human metastatic colorectal cancer tissues compared with non-metastatic ones (n = 7 per group). (C) Increased expression level of miR-4261 in human colorectal cancer tissues compared with adjacent normal tissues (n = 42 pairs). *p < 0.05. Data are presented as mean ± SEM.
miR-4261 Promotes Colorectal Cancer Cell Proliferation and Migration
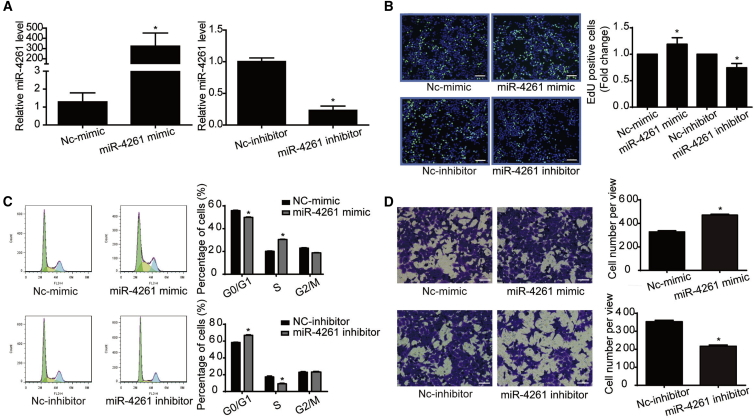
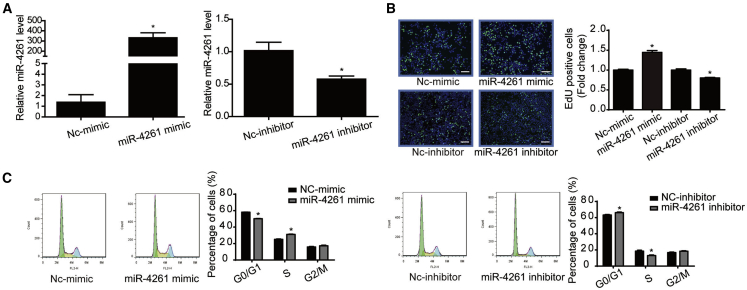
Transfection of miR-4261 mimic, inhibitor, or negative control was performed in two colorectal cancer cell lines, HCT116 and HT29, to investigate the functional roles of miR-4261 in cell proliferation and migration (Figures 2 and 3). Quantitative real-time PCR confirmed that miR-4261 mimic and miR-4261 inhibitor successfully increased or decreased miR-4261 expressions, respectively, in both HCT116 and HT29 cells (Figures 2A and 3A). After that, EdU (5-ethynyl-2-deoxyuridine) incorporation assay showed that miR-4261 mimic increased cell proliferation in both HCT116 and HT29 cell lines, while miR-4261 inhibitor reduced that (Figures 2B and 3B). Flow cytometry for cell cycle analysis showed that miR-4261 mimic accelerated the G1/S phase transition in both colorectal cancer cell lines, while the miR-4261 inhibitor had the opposite effect (Figures 2C and 3C). Moreover, transwell assay showed that miR-4261 mimic promoted, while miR-4261 inhibitor suppressed, HCT116 cell migration (Figure 2D). These data indicate that miR-4261 has a positive effect on colorectal cancer cell proliferation and migration.
Figure 2.
miR-4261 Promotes HCT116 Cell Proliferation and Migration
(A) miR-4261 mimic and inhibitor takes effect in HCT116 cells (n = 4). (B and C) miR-4261 promotes cell proliferation (B) and G1/S phase transition of cell cycle (C) in HCT116 cells, as determined by EdU incorporation assay (n = 4; scale bars, 50 μm) and flow cytometry (n = 4). EdU is represented by green and DAPI by blue. (D) miR-4261 promotes cell migration in HCT116 cells, as determined by transwell assay (n = 3, original magnification ×200). *p < 0.05. Data are presented as mean ± SEM.
Figure 3.
miR-4261 Promotes HT29 Cell Proliferation
(A) miR-4261 mimic and inhibitor takes effect in the HT29 cell line (n = 4). (B and C) miR-4261 promotes cell proliferation (B) and G1/S phase transition of cell cycle (C) in HT29 cells, as determined by EdU incorporation assay (n = 4; scale bars, 50 μm) and flow cytometry (n = 4). EdU is represented by green and DAPI by blue. *p < 0.05. Data are presented as mean ± SEM.
MCC Is a Functional Target Gene of miR-4261 in Colorectal Cancer Cells
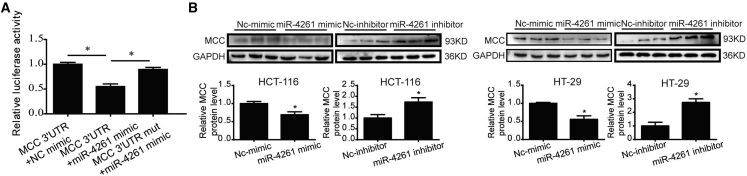
We performed miRNA target prediction algorithms based on miRDB, miRWalk, and Diana Tools, and all of them predicted MCC, a crucial tumor suppressor of colorectal cancer, as a direct target of miR-4261.25 Thus, we hypothesized that miR-4261 via targeting MCC would promote colorectal cancer. Luciferase reporter assay first validated that miR-4261 could directly target MCC, as demonstrated by reduced luciferase activity when miR-4261 combined to the 3′ UTR of MCC mRNA, while no change in luciferase activity was found when the 3′ UTR was mutated (Figure 4A). Western blot analysis further demonstrated that MCC was negatively regulated by miR-4261 in both HCT116 and HT29 cell lines (Figure 4B).
Figure 4.
miR-4261 Directly Targets MCC
(A) Luciferase reporter assay validates MCC as a direct target of miR-4261 (n = 6). (B) miR-4261 negatively regulates MCC protein level in HCT116 and HT29 cells (n = 3). *p < 0.05. Data are presented as mean ± SEM.
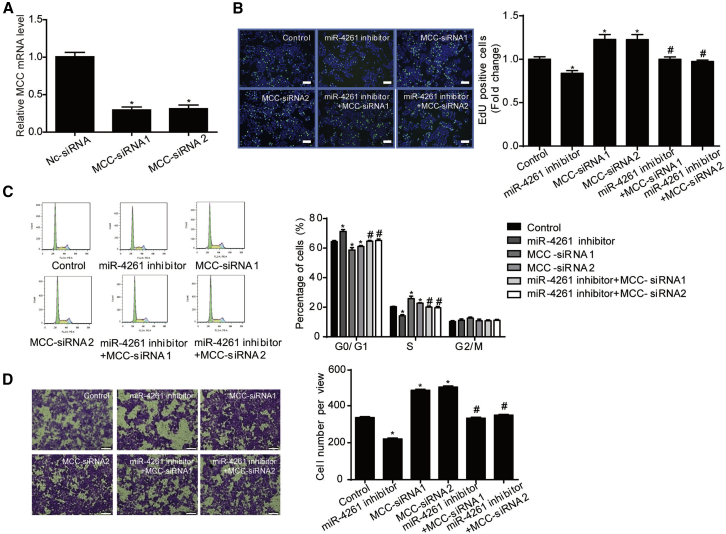
After that, HCT116 and HT29 cell lines co-transfected with MCC small interfering RNA (siRNA) and miR-4261 inhibitor were used for function-rescue assays (Figures 5 and 6). Both siRNAs used here targeting MCC were efficient to silence MCC expression in both HCT116 and HT29 cell lines (Figures 5A and 6A). Moreover, MCC siRNAs could significantly increase cell proliferation, G1/S phase transition of cell cycle, and cell migration, as well as abolish the suppressive effects of miR-4261 inhibitor on these biological processes in HCT116 (Figures 5B–5D) and HT29 cells (Figures 6B and 6C). Collectively, our results demonstrate that MCC is a functional target gene of miR-4261 in the regulation of colorectal cancer cell proliferation and migration.
Figure 5.
MCC Is a Functional Target Gene of miR-4261 in HCT116 Cells
(A) MCC siRNAs take effect in HCT116 cells (n = 4). (B–D) MCC siRNAs abolish the suppressive effect of miR-4261 inhibitor on cell proliferation, G1/S phase transition of cell cycle, and cell migration in HCT116 cells, as determined by (B) EdU incorporation assay (n = 4; scale bars, 50 μm), (C) flow cytometry (n = 4), and (D) transwell assay (n = 3; original magnification ×200). EdU is represented by green and DAPI by blue. *p < 0.05 versus control; #p < 0.05 versus miR-4261 inhibitor. Data are presented as mean ± SEM.
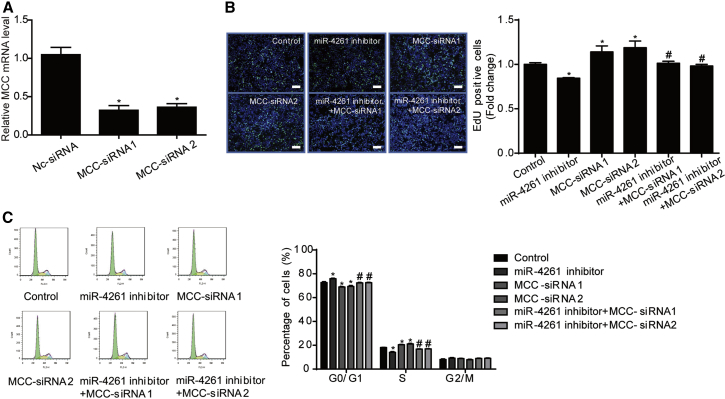
Figure 6.
MCC Is a Functional Target Gene of miR-4261 in HT29 Cells
(A) MCC siRNAs take effect in HT29 cells (n = 4). (B and C) MCC siRNAs abolish the suppressive effect of the miR-4261 inhibitor on cell proliferation (B) and G1/S phase transition of the cell cycle (C) in HT29 cells, as determined by EdU incorporation assay (n = 4; scale bars, 50 μm) and flow cytometry (n = 4). EdU is represented by green and DAPI by blue. *p < 0.05 versus control; #p < 0.05 versus miR-4261 inhibitor. Data are presented as mean ± SEM.
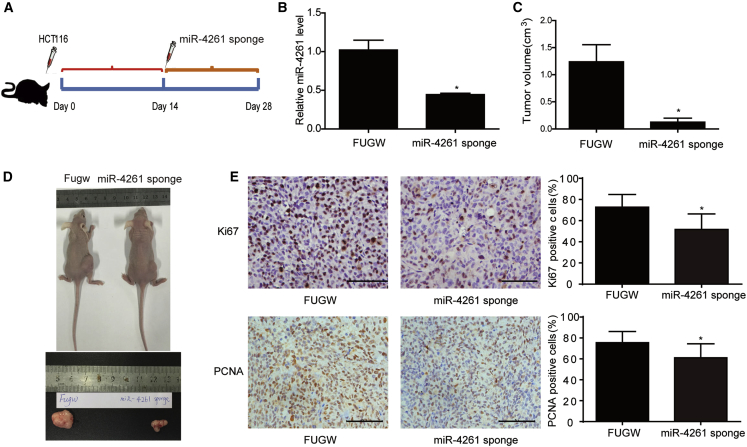
miR-4261 Suppression Has a Therapeutic Effect on Colorectal Cancer
Lentivirus-based miR-4261 sponge was constructed and subcutaneously injected around the xenograft 14 days after mice were implanted with HCT116 cells (Figure 7A). We first validated the efficiency of miR-4261 sponge as demonstrated by significant suppression of miR-4261 expression level in the xenograft harvested on day 28 (14 days after miR-4261 sponge injection) (Figure 7B). Importantly, miR-4261 sponge was efficient to inhibit tumor growth (Figures 7C and 7D). Immunohistochemical analysis for Ki67 and proliferating cell nuclear antigen (PCNA) further indicated that miR-4261 sponge also reduced cell proliferation in the colorectal cancer xenograft (Figure 7E). These data provide in vivo evidence that miR-4261 inhibition has a therapeutic effect in colorectal cancer.
Figure 7.
Therapeutic Effect of miR-4261 Suppression on Colorectal Cancer
(A) Schema for subcutaneous injection of miR-4261 sponge 14 days after mice were implanted with HCT116 cells (n = 5). (B) Reduced miR-4261 expression in the xenograft with miR-4261 sponge treatment (n = 4). (C and D) Reduced tumor volume (n = 5) in mice treated with miR-4261 sponge. (E) Reduced cell proliferation in the colorectal cancer xenograft after miR-4261 sponge treatment, as determined by Ki67 and PCNA immunohistochemistry (n = 5; original magnification ×200). *p < 0.05. Data are presented as mean ± SEM.
Discussion
Increasing studies have demonstrated the miRNA-mRNA interactions in colorectal cancer by emphasizing their regulatory effects on different cellular processes such as proliferation, apoptosis, migration, invasion, and angiogenesis.10, 12, 18, 29 However, among the increasing number of specific target genes identified for dysregulated miRNAs in colorectal cancer, few studies have reported the miRNA(s) that could directly target MCC.6 MCC has recently been demonstrated to be targeted by miR-4260, which contributes to the development of colorectal cancer.30 Because the silence or downregulation of the tumor suppressor gene MCC has been considered as an early event in colorectal carcinogenesis,22, 25 identifying the miRNA(s) that could inhibit MCC may act as an important contributor(s) to colorectal cancer.
In the present study, we applied the miRNA target prediction algorithms by using miRDB, miRWalk, and Diana Tools databases, and referred to the miRNA microarray data in human colorectal cancer specimens previously published and registered to GEO (accession number GEO: GSE72199).27 Among the predicted miRNAs that could target MCC, only miR-4261 was reported to be dysregulated in colorectal cancer, as indicated in the microarray data.27 Importantly, we further validated that miR-4261 was overexpressed in metastatic colorectal cancer tissues compared with non-metastatic ones, and also upregulated in colorectal cancer tissues compared with normal adjacent tissues based on our own cohort of patients clinically diagnosed as colorectal cancer at the Department of General Surgery of Tongji Hospital. Our investigations on the biological roles of miR-4261 in colorectal cancer further revealed that overexpression of miR-4261 led to the promotion of cell proliferation, cell cycle progression from G0/G1 to S phase, and cell migration in HCT116 and HT29 cell lines, and suppression of miR-4261 was able to inhibit these cellular processes. Excitedly, to date, no functional effect of miR-4261 has been reported in any disease processes, and our present findings might provide first insight into the potential roles of miR-4261 in colorectal cancer through promotion of cancer cell proliferation and migration.
MCC is an important colorectal tumor suppressor gene whose downregulation can be an early event in colorectal carcinogenesis.22, 25 MCC has a negative regulatory effect on cell growth by blocking G1/S phase transition in mouse NIH 3T3 cells.24 The inhibitory effect of MCC on colorectal cancer cell proliferation was found to be mediated by inhibition of the Wnt/β-catenin signaling.25, 26 In addition, MCC regulates cell migration in epithelial cells31 and also acts as a transcriptional regulator of nuclear factor κB (NF-κB) in colorectal cancer cells.32 Therefore, MCC may have multiple biological cellular functions during the development of colorectal cancer. Here, luciferase reporter assay and western blot analysis first validated that miR-4261 could directly target the 3′ UTR of MCC mRNA, and negatively regulate MCC protein level in HCT116 and HT29 cell lines. Function-rescue assays were further performed to identify MCC as a functional target gene of miR-4261 involved in the cell proliferation and migration of colorectal cancer cells. Indeed, our data provide direct evidence that miR-4261 through targeting of MCC promotes colorectal cancer.
miRNA-based therapeutics or inhibition of overexpressed oncogenic miRNAs might be useful therapeutic interventions for cancers.33, 34, 35, 36 Based on our data from in vitro experiments, we further investigated the potential of miR-4261 inhibition as a molecularly targeted therapy in colorectal cancer. Importantly, therapeutic intervention with lentivirus-based miR-4261 sponge injection was quite effective to reduce tumor growth and inhibit cell proliferation even in the established colorectal cancer xenograft, which indicates that inhibition of miR-4261 may serve as a potential therapeutic strategy for colorectal cancer. In this case, further studies are required to clarify the upstream mechanisms inducing the dysregulation of miR-4261 during colorectal cancer carcinogenesis. It is also important to identify other target genes of miR-4261 that might be involved in the regulation of colorectal cancer.
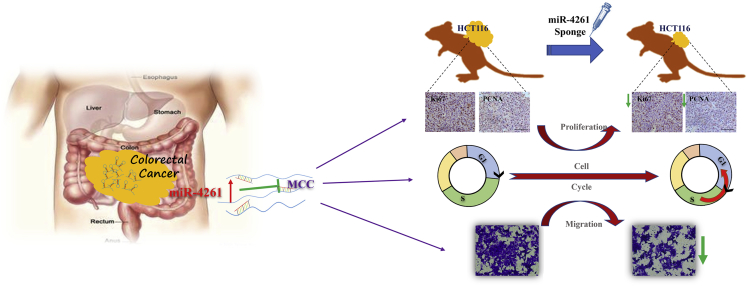
In summary, our study identifies miR-4261 as an upstream regulator of MCC and further demonstrates the oncogenic effect of miR-4261 in colorectal cancer via promoting cancer cell proliferation and migration, while inhibition of miR-4261 is effective to prevent colorectal cancer development (Figure 8). Our study is the first to unravel the functional role of miR-4261, and it provides strong evidence that inhibition of miR-4261 through targeting of MCC might exert a therapeutic effect for colorectal cancer.
Figure 8.
Schematic Diagram Showing That Inhibition of miR-4261 through Targeting of MCC Prevents Tumor Cell Proliferation and Migration and Inhibits Colorectal Cancer Growth
Materials and Methods
Colorectal Cancer Cell Culture and Transfection
Two human colorectal cancer cell lines HT29 and HCT116 were respectively cultured in RPMI 1640 (KeyGen Biotech) and DMEM (Corning) supplemented with 1% penicillin and streptomycin (GIBCO) and 10% fetal bovine serum (FBS; HyClone) at 37°C in a 5% CO2 atmosphere. To investigate the functional roles of miR-4261, we seeded HT29 and HCT116 cells (2 × 105/mL) on plates, starved them for 6 hr, and then transfected them with miR-4261 mimic (50 nM; RiboBio), inhibitor (100 nM; RiboBio), or negative control for 48 hr using Lipofectamine 2000 (Invitrogen). To determine whether MCC is a functional target of miR-4261, we performed function-rescue assays in HT29 and HCT116 cells co-transfected with MCC siRNA (75 nM; RiboBio) and miR-4261 inhibitor (100 nM; RiboBio) for 48 hr using Lipofectamine 2000.
Animal Experiment of Colorectal Cancer Xenograft
Ethical approval for tissue collection protocol was obtained from the research ethics committee of Shanghai Tongji Hospital. The protocol for this study was approved by the Animal Experiments Ethics Committee of School of Life Science at the Shanghai University. Eight-week-old BALB/c nude mice were purchased from CAVENS Lab Animal and maintained under special pathogen-free conditions. A total of 3 × 106 HCT116 cells in log phase were subcutaneously implanted to the right flank of nude mice on day 0 to induce colorectal cancer xenograft. Tumor volumes (π/6 × smaller diameter2 × larger diameter) were measured by caliper. Fourteen days after xenograft implantation (day 14), mice were randomly assigned to two groups: treated with lentivirus-based miR-4261 sponge (treatment group) or vehicle alone (Fugw control group). In brief, the sequences of miR-4261 sponge were designed and inserted into the Fugw. 293T cells were co-transfected for 48–72 hr with psPAX2, pMD2.G, and Fugw-miR-4261 sponge at the ratio of 3:1:4 using FuGene Transfection Reagent (Roche). After that, the medium was harvested, centrifugated, and filtrated for lentivirus collection. For the treatment group, 50 μL of lentivirus-based miR-4261 sponge at 108 plaque-forming units (PFUs) was subcutaneously injected around the xenograft on day 14. Mice in the control group were injected with Fugw control. Finally, mice were sacrificed and the xenograft tissues were harvested after another 14 days (day 28).
Tissue Samples from Colorectal Cancer Patients
A total of 42 pairs of colorectal cancer tissues versus adjacent normal tissues were collected from colorectal cancer patients with informed consent at the Department of General Surgery of Tongji Hospital. The tumor tissue underwent macro-dissection to enhance the tumor content of the study material. Additionally, we collected seven metastatic versus seven non-metastatic colorectal cancer tissues, and their clinical characteristics were listed in Table 1. Human tissue samples were freshly kept in liquid nitrogen and stored at −80°C before RNA extraction.
Table 1.
The Clinical Characteristics of Colorectal Cancer Patients with or without Liver Metastasis
| Non-metastatic (n = 7) | Metastatic (n = 7) | |
|---|---|---|
| Age (X ± S, years) | 62 ± 5 | 62 ± 8 |
| Gender (n) | ||
| Male | 5 | 5 |
| Female | 2 | 2 |
| Heart rate (X ± S, beats/min) | 79 ± 6 | 76 ± 7 |
| BMI (X ± S, kg/m2) | 23.5 ± 0.8 | 24.6 ± 1.2 |
| Depth (n) | ||
| T1, T2 | 2 | 0 |
| T3, T4 | 5 | 7 |
| Lymph node (n) | ||
| N0, N1 | 6 | 3 |
| N2 | 1 | 4 |
| Tumor location (n) | ||
| Colon | 6 | 3 |
| Rectum | 1 | 4 |
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from tissues or cells using TRIzol (TaKaRa) according to the manufacturer’s instructions. The final concentration of RNA samples was determined by NanoDrop mass spectrometry (Thermo). For MCC mRNA analysis, first-strand cDNA was obtained using the cDNA reverse transcription kit (TaKaRa), and the SYBR (TaKaRa) was used for qPCR on the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). GAPDH was used as internal control. The primer sequences for MCC and GAPDH were as follows: MCC: forward 5′-AATAAACGTCTCCAGCAAACAGA-3′, reverse 5′-CGTTCCTCATAGCGAAGTGTC-3′; GAPDH: forward 5′-ATGACATCAAGAAGGTGGTG-3′, reverse 5′-CATACCAGGAAATG AGCTTG-3′. For miRNA analysis, total RNA was reverse transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). The expression level of miR-4261 was determined using the Bulge-Loop miRNA qPCR Primer Set (RiboBio) with SYBR (TaKaRa) on ABI 7900HT Fast Real-Time PCR System. 5 s was used as internal control.
Western Blotting
Total protein was extracted from cells using radioimmunoprecipitation (RIPA) lysis buffer (Beyotime) supplemented with 1% PMSF (KeyGen Biotech). Equal quantity of total protein samples was separated on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes. Blots were incubated overnight at 4°C with primary anti-MCC antibody (1:1,000; Abcam). When secondary antibody incubation finished, the electrochemiluminescence (ECL) kit (Tanon) was used to visualize protein signals. GAPDH was used as internal control.
EdU Incorporation Assay
To investigate the role of miR-4261 in cell proliferation, we seeded HCT116 and HT29 cells at a density of 2 × 105/mL in 96-well plates and transfected them with miR-4261 mimic, inhibitor, or negative control for 48 hr. Cells were then incubated with EdU for 2 hr before the end of the experiment and stained according to the instructions of EdU assay (Ribobio). Nuclei were counterstained with DAPI. EdU- and DAPI-positive cells were observed under a fluorescent microscope (Leica). The proportion of EdU-positive cells to total cells was analyzed by ImageJ software.
Flow Cytometry for Cell Cycle
After transfection for 48 hr, HCT116 and HT29 cells were fixed with cold absolute ethanol overnight. Cells were subsequently treated with RNase A (KeyGen Biotech) and stained with propidium iodide (PI; Sigma) in the dark at room temperature according to the manufacturer’s protocol. Cell cycle was determined by flow cytometry (Beckman). The proportion of cells in G0/G1, S, and G2/M phases was calculated using FlowJo software.
Transwell Assay
Cell migration was determined by transwell system with polycarbonate membrane (pore size 8.0 μm; Corning). In brief, HCT116 cells were first transfected with miR-4261 mimic, inhibitor, or negative control for 24 hr and resuspended in 200 μL of FBS-free DMEM. Cells were then seeded at a density of 2 × 105/mL in the upper chamber of insert, while the lower chamber was filled with 600 μL of DMEM containing 10% FBS. After 48 hr, cells in the upper chamber were erased with cotton, and the passing cells in the lower layer were fixed with 4% paraformaldehyde (PFA) and stained with crystal violet (Beyotime). Penetrating cells were observed under an inverted research microscope (Leica).
Immunohistochemical Staining
The colorectal cancer xenograft tissues were fixed with 4% PFA and then paraffined. Five-micrometer-thick sections were deparaffinized with xylene and treated with decreasing concentrations of ethanol for hydration. Antigen retrieval was carried out by high-pressure boiling of tissue sections in sodium citrate buffer at pH 6.0. Endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide in methanol. After that, tissue sections were blocked with 5% BSA and incubated with primary anti-PCNA (1:300; Abcam) and anti-Ki67 (1:300; Abcam) antibodies overnight at 4°C in a humidified chamber. After incubation with biotinylated secondary antibody, immunohistochemical stainings for PCNA and Ki67 were developed with the 3,3′-diaminobenzidine tetrahydrochloride (DAB) substrate chromogen system (KeyGen Biotech). Finally, hematoxylin was added as a counterstain, and tissue sections were dehydrated in increasing concentrations of ethanol. The PCNA- and Ki67-positive cells were photographed with an inverted research microscope (Leica).
Luciferase Reporter Assay
The miRDB database was used to predict the miR-4261 binding site on the 3′ UTR of MCC mRNA. A fragment of the 3′ UTR of MCC mRNA containing the putative miR-4261 binding sequence was obtained by PCR amplification and then cloned into the pGL3-Basic Vector (Promega). The primers used were listed in Table 2. Luciferase reporter assay was performed using a dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. In brief, 293T cells were co-transfected with pGL3-basic-3′ UTR or pGL3-basic-3′ UTR mut, Renilla, and miR-4261 inhibitor or negative control for 24 hr using Lipofectamine 2000 (Invitrogen). Luciferase activities were finally measured using a plate-reading luminometer.
Table 2.
The Primer Sequences Used in the Luciferase Reporter Assay
| Primer | Sequences |
|---|---|
| MCC 3′ UTR | normal, forward: 5′-GGGTCTAGATCCCTTCTGAAGGTGTTTCC T-3′ |
| mutant, forward: 5′-GGGTCTAGATCCCTTCTGAAGGACAAAGGT-3′ | |
| reverse: 5′-GGGTCTAGAGCTGAATCTAGAAGTAGGTAGA-3′ |
The underlined text is the seed sequence used in the luciferase reporter assay.
Statistical Analysis
Data were presented as mean ± SEM. All statistical analyses were performed using SPSS 20.0 software. For cell and animal experiments, intergroup comparisons were carried out by independent-samples t test or one-way ANOVA test. For human specimens, paired-samples t test was used to compare miR-4261 expression level in colorectal cancer tissues versus adjacent normal tissues. A p value < 0.05 was considered statistically significant.
Author Contributions
J.X. and J.Z. designed the study, supervised all experiments, drafted the manuscript, and contributed equally to this work. G.J., Q.H., and M.H. performed the experiments and contributed equally to this work. X.L., F.L., C.L., W.F., Y.A., and B.X. helped perform the experiments and analyzed the data.
Conflicts of Interest
The authors declare there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81570362, 81200169, and 91639101 to J.X. and grant 21672136 to B.X.) and the development fund for Shanghai talents (to J.X.).
Contributor Information
Jinzhe Zhou, Email: zhoujinzhe_@126.com.
Junjie Xiao, Email: junjiexiao@live.cn.
References
- 1.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.De Sousa E Melo F., Wang X., Jansen M., Fessler E., Trinh A., de Rooij L.P., de Jong J.H., de Boer O.J., van Leersum R., Bijlsma M.F. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 3.Sadanandam A., Lyssiotis C.A., Homicsko K., Collisson E.A., Gibb W.J., Wullschleger S., Ostos L.C., Lannon W.A., Grotzinger C., Del Rio M. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold D., Seufferlein T. Targeted treatments in colorectal cancer: state of the art and future perspectives. Gut. 2010;59:838–858. doi: 10.1136/gut.2009.196006. [DOI] [PubMed] [Google Scholar]
- 5.van Gijn W., Marijnen C.A., Nagtegaal I.D., Kranenbarg E.M., Putter H., Wiggers T., Rutten H.J., Påhlman L., Glimelius B., van de Velde C.J., Dutch Colorectal Cancer Group Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 6.Amirkhah R., Schmitz U., Linnebacher M., Wolkenhauer O., Farazmand A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54:129–141. doi: 10.1002/gcc.22231. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y., Zhang P., Wang F., Zhang H., Yang Y., Shi C., Xia Y., Peng J., Liu W., Yang Z., Qin H. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat. Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 8.Sun J.Y., Huang Y., Li J.P., Zhang X., Wang L., Meng Y.L., Yan B., Bian Y.Q., Zhao J., Wang W.Z. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem. Biophys. Res. Commun. 2012;420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 10.Xiong B., Cheng Y., Ma L., Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int. J. Oncol. 2013;42:219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- 11.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Chen L., Xu Y., Li R., Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 13.Nie J., Liu L., Zheng W., Chen L., Wu X., Xu Y., Du X., Han W. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2012;33:220–225. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- 14.Yin Y., Yan Z.P., Lu N.N., Xu Q., He J., Qian X., Yu J., Guan X., Jiang B.H., Liu L.Z. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta. 2013;1829:239–247. doi: 10.1016/j.bbagrm.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Xu Q., Liu L.Z., Qian X., Chen Q., Jiang Y., Li D., Lai L., Jiang B.H. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761–774. doi: 10.1093/nar/gkr730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H.Y., Lin Y.M., Chung H.C., Lang Y.D., Lin C.J., Huang J., Wang W.C., Lin F.M., Chen Z., Huang H.D. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 17.Hur K., Toiyama Y., Takahashi M., Balaguer F., Nagasaka T., Koike J., Hemmi H., Koi M., Boland C.R., Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J., Luo M. MicroRNA-221 promotes colorectal cancer cell invasion and metastasis by targeting RECK. FEBS Lett. 2014;588:99–104. doi: 10.1016/j.febslet.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Nishisho I., Nakamura Y., Miyoshi Y., Miki Y., Ando H., Horii A., Koyama K., Utsunomiya J., Baba S., Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 20.Kinzler K.W., Nilbert M.C., Vogelstein B., Bryan T.M., Levy D.B., Smith K.J., Preisinger A.C., Hamilton S.R., Hedge P., Markham A. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366–1370. doi: 10.1126/science.1848370. [DOI] [PubMed] [Google Scholar]
- 21.Starr T.K., Allaei R., Silverstein K.A., Staggs R.A., Sarver A.L., Bergemann T.L., Gupta M., O’Sullivan M.G., Matise I., Dupuy A.J. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohonen-Corish M.R., Sigglekow N.D., Susanto J., Chapuis P.H., Bokey E.L., Dent O.F., Chan C., Lin B.P., Seng T.J., Laird P.W. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–4441. doi: 10.1038/sj.onc.1210210. [DOI] [PubMed] [Google Scholar]
- 23.Ashton-Rickardt P.G., Wyllie A.H., Bird C.C., Dunlop M.G., Steel C.M., Morris R.G., Piris J., Romanowski P., Wood R., White R. MCC, a candidate familial polyposis gene in 5q.21, shows frequent allele loss in colorectal and lung cancer. Oncogene. 1991;6:1881–1886. [PubMed] [Google Scholar]
- 24.Matsumine A., Senda T., Baeg G.H., Roy B.C., Nakamura Y., Noda M., Toyoshima K., Akiyama T. MCC, a cytoplasmic protein that blocks cell cycle progression from the G0/G1 to S phase. J. Biol. Chem. 1996;271:10341–10346. doi: 10.1074/jbc.271.17.10341. [DOI] [PubMed] [Google Scholar]
- 25.Fukuyama R., Niculaita R., Ng K.P., Obusez E., Sanchez J., Kalady M., Aung P.P., Casey G., Sizemore N. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene. 2008;27:6044–6055. doi: 10.1038/onc.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pangon L., Mladenova D., Watkins L., Van Kralingen C., Currey N., Al-Sohaily S., Lecine P., Borg J.P., Kohonen-Corish M.R. MCC inhibits beta-catenin transcriptional activity by sequestering DBC1 in the cytoplasm. Int. J. Cancer. 2015;136:55–64. doi: 10.1002/ijc.28967. [DOI] [PubMed] [Google Scholar]
- 27.Hata T., Mokutani Y., Takahashi H., Inoue A., Munakata K., Nagata K., Haraguchi N., Nishimura J., Hata T., Matsuda C. Identification of microRNA-487b as a negative regulator of liver metastasis by regulation of KRAS in colorectal cancer. Int. J. Oncol. 2017;50:487–496. doi: 10.3892/ijo.2016.3813. [DOI] [PubMed] [Google Scholar]
- 28.Goff L.A., Davila J., Swerdel M.R., Moore J.C., Cohen R.I., Wu H., Sun Y.E., Hart R.P. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huangfu L., Liang H., Wang G., Su X., Li L., Du Z., Hu M., Dong Y., Bai X., Liu T. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget. 2016;7:4735–4745. doi: 10.18632/oncotarget.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao J., Lv D., Zhou J., Bei Y., Chen T., Hu M., Zhou Q., Fu S., Huang Q. Therapeutic inhibition of miR-4260 suppresses colorectal cancer via targeting MCC and SMAD4. Theranostics. 2017;7:1901–1913. doi: 10.7150/thno.19168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnaud C., Sebbagh M., Nola S., Audebert S., Bidaut G., Hermant A., Gayet O., Dusetti N.J., Ollendorff V., Santoni M.J. MCC, a new interacting protein for Scrib, is required for cell migration in epithelial cells. FEBS Lett. 2009;583:2326–2332. doi: 10.1016/j.febslet.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 32.Sigglekow N.D., Pangon L., Brummer T., Molloy M., Hawkins N.J., Ward R.L., Musgrove E.A., Kohonen-Corish M.R. Mutated in colorectal cancer protein modulates the NFκB pathway. Anticancer Res. 2012;32:73–79. [PubMed] [Google Scholar]
- 33.Li Q., Zou C., Zou C., Han Z., Xiao H., Wei H., Wang W., Zhang L., Zhang X., Tang Q. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335:168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 34.Bao Y., Chen Z., Guo Y., Feng Y., Li Z., Han W., Wang J., Zhao W., Jiao Y., Li K. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS ONE. 2014;9:e105991. doi: 10.1371/journal.pone.0105991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng B., Dong T.T., Wang L.L., Zhou H.M., Zhao H.C., Dong F., Zheng M.H. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS ONE. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M., Tang Q., Qiu M., Lang N., Li M., Zheng Y., Bi F. miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett. 2011;585:2998–3005. doi: 10.1016/j.febslet.2011.08.014. [DOI] [PubMed] [Google Scholar]