Figure S2.
Structure Determination and Functional Analysis of LysinR, Related to Figure 2
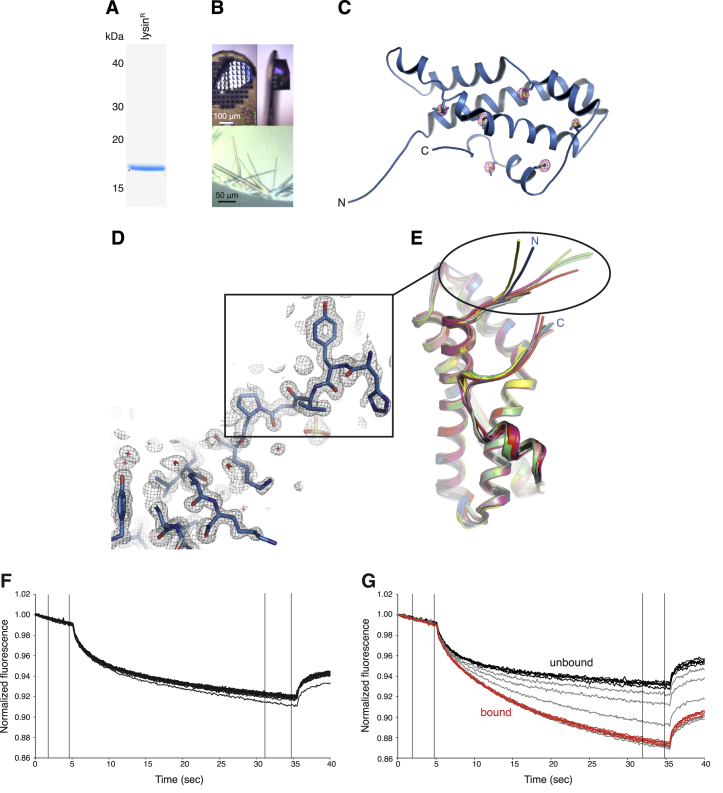
(A) Coomassie-stained SDS-PAGE analysis of purified lysinR.
(B) Orthorhombic (top panels) and monoclinic (bottom panel) crystals of lysinR.
(C) The sulfur atoms of 6 Met residues are identified by a 5 σ anomalous difference map of lysinR (red mesh), which is shown superimposed onto the refined model of the protein.
(D) Detail of the hypervariable N-terminal region of lysinR in the refined 0.99 Å resolution 2mFo-DFc electron density map of the protein, contoured at 1 σ.
(E) Comparison of chains from different structures highlights the flexibility of lysin N terminus. Blue, orthorhombic lysinR; shades of green, chains of monoclinic lysinR; shades of gray, chains of PDB: 1LYN; yellow, PDB: 2LIS; shades of red, chains of PDB: 2LYN.
(F and G) MST analysis of the interaction of lysinR with different VERL repeats. MST time traces obtained by titrating 10 nM labeled lysin with increasing concentrations of VR1+ up to 0.1 mM (F) and VR3 up to 2 μM (G). Traces corresponding to bound and unbound states are colored red and black, respectively, whereas traces corresponding to partially bound intermediates are shown in gray. VR3 binds to lysin with high affinity, but no binding is detected to VR1+.