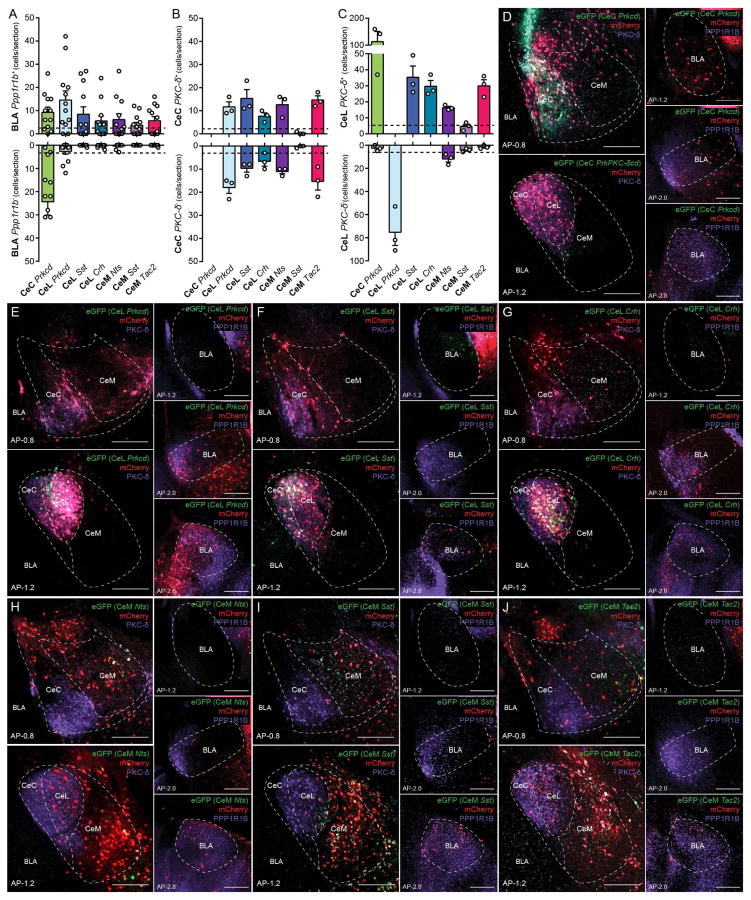
Figure 5. Monosynaptic retrograde tracing from genetically defined CeA neurons.
(A) Quantification of rabies-mediated retrograde labeled PPP1R1B+ and PPP1R1B − neurons in the BLA from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per section in the BLA from n = 8 sections per mouse from n = 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 2.5 neurons per section was used to construct a connectivity model (Figure 8A).
(B) Quantification of rabies-mediated retrograde labeled PKC-δ+ and PKC-δ − neurons in the CeC from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per single section in the CeC from n = 3 sections from 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 2.5 neurons per section was used to construct a connectivity model (Figure 8A).
(C) Quantification of rabies-mediated retrograde labeled PKC-δ + and PKC-δ − neurons in the CeL from genetically defined CeA neurons. Individual points represent the number of retrograde labeled neurons per single section in the CeL from n = 3 sections from 3 mice, bars represent mean ± s.e.m.. Arbitrary threshold of 5 neurons per section was used to construct a connectivity model (Figure 8A).
(D–J) Representative histology of rabies-mediated retrograde tracing from CeC Prkcd+ neurons (D), CeL Prkcd+ neurons (E), CeL Sst+ neurons (F), CeL Crh+Nts+Tac2+ neurons (G), CeM Nts+ neurons (H), CeM Sst+ neurons (I), CeM Tac2+ neurons (J) in the CeA and BLA. Starter cells are labeled with eGFP (green) and mCherry (red) and mouse line of starter cells are noted (italicized), Rabies virus is labeled with mCherry (red). Prkcd protein (purple) was labeled in the CeA. Ppp1r1b protein (purple) was labeled in BLA. The anterior-posterior (AP) distance from Bregma (mm), scale bar, 250 μm.