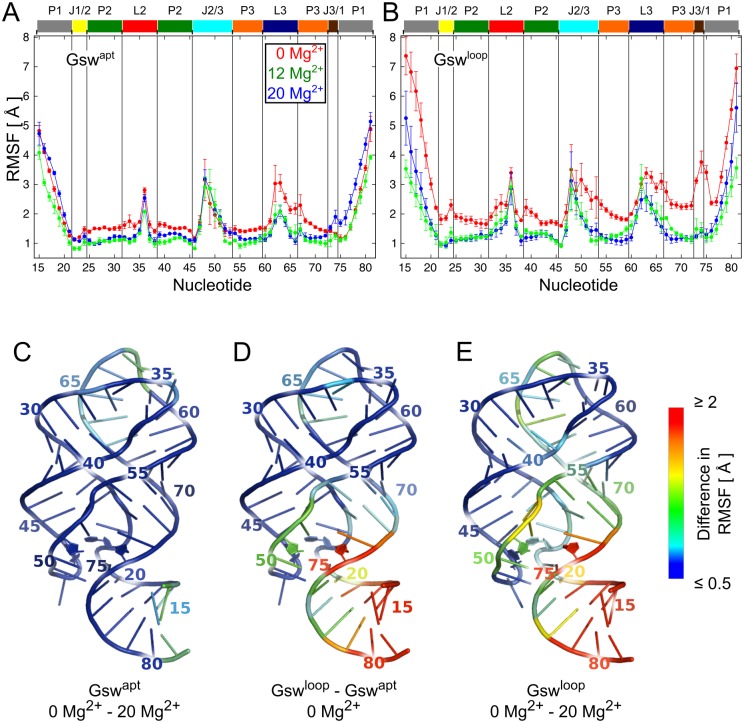
Fig 2. Atomic fluctuations calculated from MD simulations.
A, B: Mean (± SEM) atomic fluctuations (RMSF) on a per nucleotide level for Gswapt (A) and Gswloop (B) over the three simulations for each system setup. Secondary structure regions are depicted above the plots and colored according to Fig 1A. Red: simulations in the absence of Mg2+ ions; green: simulations with 12 Mg2+ ions per RNA molecule; blue: simulations with 20 Mg2+ ions per RNA molecule. C, D, E: Differences in atomic fluctuations projected onto the RNA structure. Larger differences are colored red, smaller differences blue. Nucleotides responsible for ligand binding are shown as sticks. C: Difference in atomic fluctuations for Gswapt of 0 Mg2+—20 Mg2+ ions per RNA molecule. D: Difference in atomic fluctuations in the absence of Mg2+ for Gswloop—Gswapt. E: Difference in atomic fluctuations for Gswloop of 0 Mg2+—20 Mg2+ ions per RNA molecule.