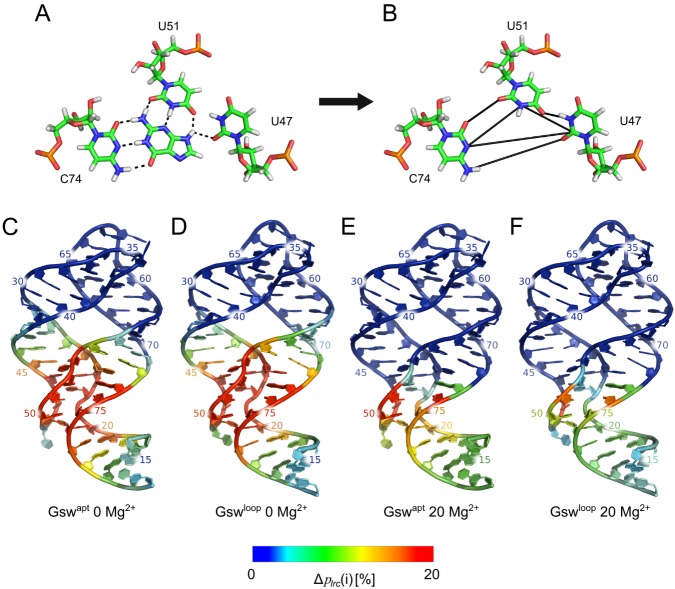
Fig 3. Rigidity analyses.
A: Nucleotides involved in binding guanine in the binding site of the aptamer domain of Gsw (PDB ID 1Y27 [26]). Black dashed lines indicate hydrogen bonds. B: Constraints (black lines) added to conformations of the apo aptamer domain of Gsw to model the presence of guanine in the binding site for rigidity analyses. C, D, E, F: Nucleotide-wise differences in the probability to be in the largest rigid cluster (Δplrc(i), Eq 1, EHB = −0.6 kcal/mol for the rigidity analyses) between the ligand being present or absent in the aptamer domain, projected onto the aptamer domain of Gswapt (C, E) and Gswloop (D, F) in the absence of Mg2+ ions (C, D) and in the presence of 20 Mg2+ ions (E, F).