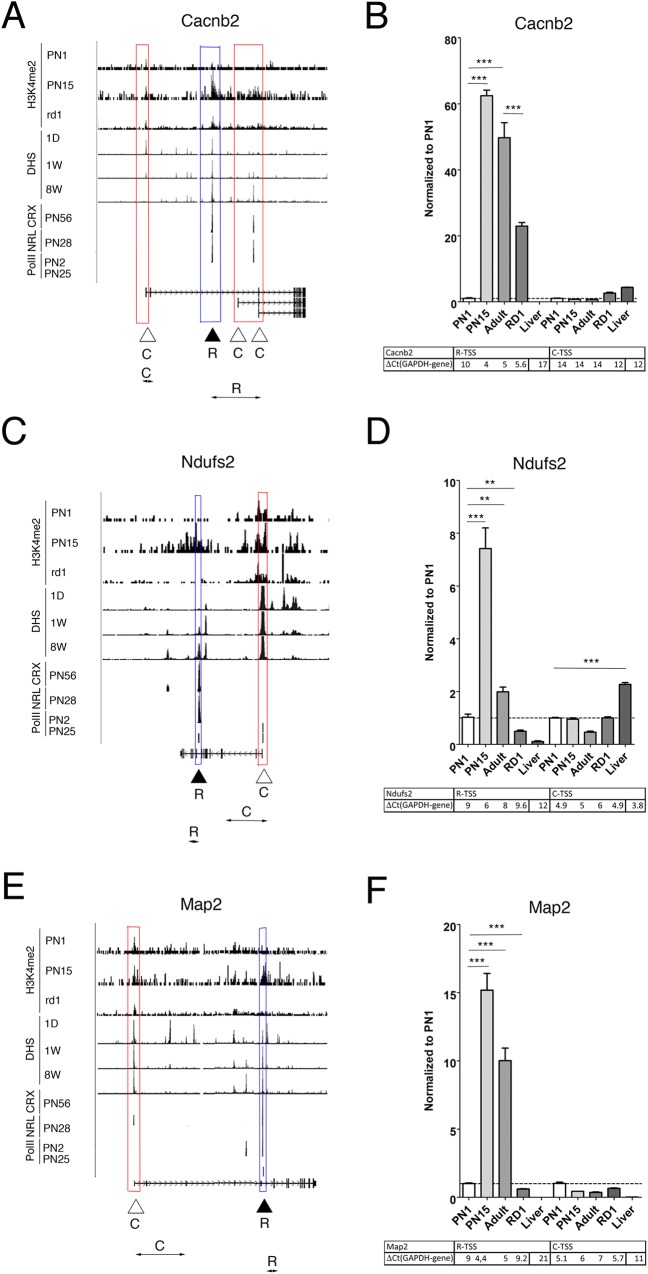
Fig 9. Chromatin features predict tissue specific new TSS.
A, C, E. Combined genome-wide tracks of chromatin features, as in Fig 1B for Cacnb2 (A), Ndufs2 (C) and Map2 (E) genes. Common TSS is depicted as C/ red box, predicted rod TSS–as R/ blue box. Position of the specific primer sets and PCR product that were used to assess and distinguish TSS-specific gene expression by RT-PCR depicted as double arrow below gene maps. B, D, F. Relative gene expressions from predicted rod and common TSS by RT-PCR with primer pairs depicted at A, C, E for Cacnb2 (B), Ndufs2 (D) and Map2 (F) for mouse retina samples at PN1, PN15, adult and RD1 mutant, compare with mouse liver. For comparison, normalized to Gapdh ΔCt values for each sample are in table below. Experiments done in duplicates with three technical replicas; ***—p < 0.0001, **—p < 0.001.