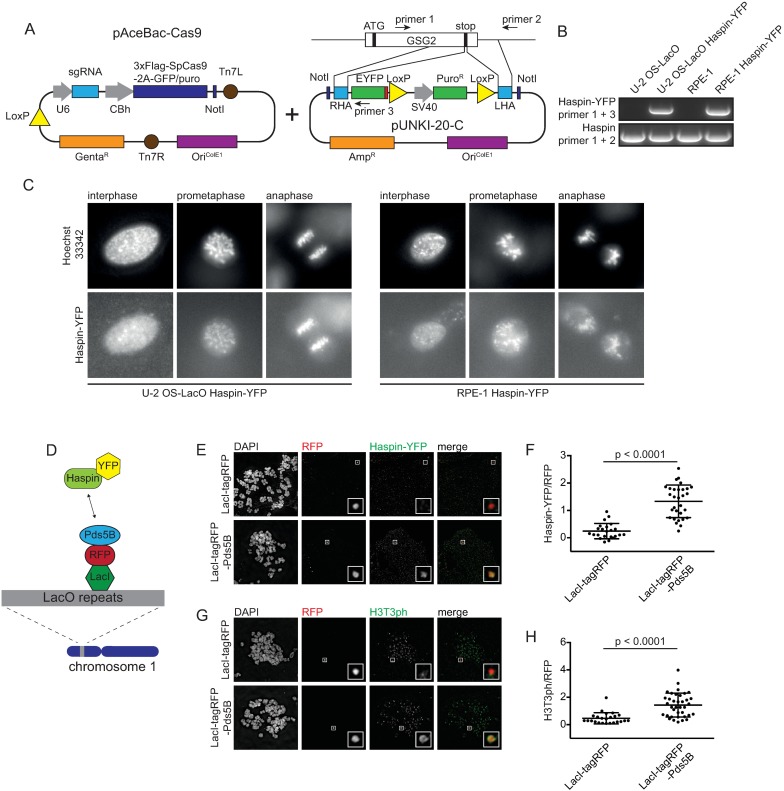
Fig 5. Endogenous tagging of Haspin using CRISPR/Cas9 baculovirus.
A) Schematic representation of the introduction of an HDR template for endogenous tagging of GSG2 in the pAceBac-Cas9 plasmid. B) Integration of the YFP-tag at the C-terminus of the gene encoding Haspin was confirmed by PCR using the indicated primer pairs, schematically depicted in Fig 5A. The PCR product obtained using primers 1 and 2 is of the untagged allele, based on its size. C) Representative live cell images of U-2 OS-LacO Haspin-YFP cells and RPE-1 Haspin-YFP cells in prometaphase and interphase. D) Schematic overview depicting the binding of LacI-tagRFP-Pds5B to the LacO repeats. Upon interaction between Pds5B and Haspin an YFP signal can be detected at this ectopic locus. E) Immunofluorescence images of metaphase spreads of U-2 OS-LacO Haspin-YFP cells expressing LacI-tagRFP or LacI-tagRFP-Pds5B and stained for DAPI, YFP and RFP. F) Quantification of Haspin-YFP at the LacO locus. Depicted is the mean of Haspin-YFP normalized over RFP ± SD. Each dot represents a single cell. The data was analyzed using an un-paired Student’s t-test. G) Immunofluorescence images of metaphase spreads of U-2 OS-LacO Haspin-YFP cells expressing LacI-tagRFP or LacI-tagRFP-Pds5B stained for DAPI, H3T3ph and RFP. H) Quantifications of H3T3ph at the LacO locus. Depicted is the mean of H3T3ph levels normalized over RFP ± the SD. Each dot represents a single cell. The data was analyzed using an un-paired Student’s t-test. A minimum of 23 cells was analyzed per experiment.