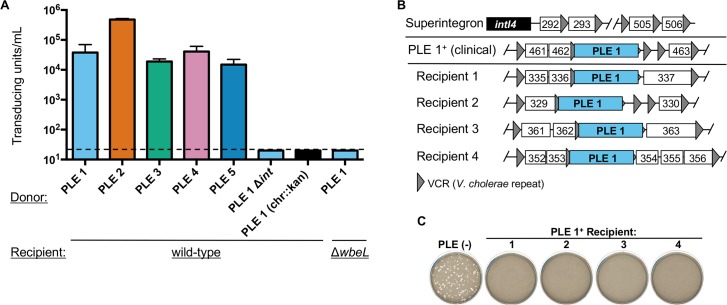
Fig 6. PLEs are mobilized following infection by ICP1-related phages.
(A) PLE transducing units produced during infection with ICP1_2006_E ΔCRISPR. When the donor strain was a PLE variant harboring the kanamycin resistance cassette elsewhere in the chromosome (designated as chr::kan), no transduction could be detected, but we included only the PLE 1 variant for simplicity. The dashed line indicates the limit of detection for this assay. (B) PLE 1 integration into the V. cholerae superintegron. The V. cholerae superintegron is schematized in the top row: the superintegron integrase gene (intI4) with proximal and distal ORFs defining the superintegron boundaries are shown. ORFs are indicated by white boxes with 3 digit numbers corresponding to the VCA0XXX designation as observed in the N16961 reference genome. The position of PLE 1 in clinical isolates and four recipients generated by ICP1-mediated transduction is indicated by the PLE flanking genes within the superintegron. (C) Experimental PLE 1 transductants show resistance to ICP1 regardless of the position of PLE 1 within the superintegron. The sensitivity of each of the four PLE 1 recipients in (B) to ICP1_2011_A lacking CRISPR is shown. For panel A, error bars indicate standard deviations of biological triplicates.