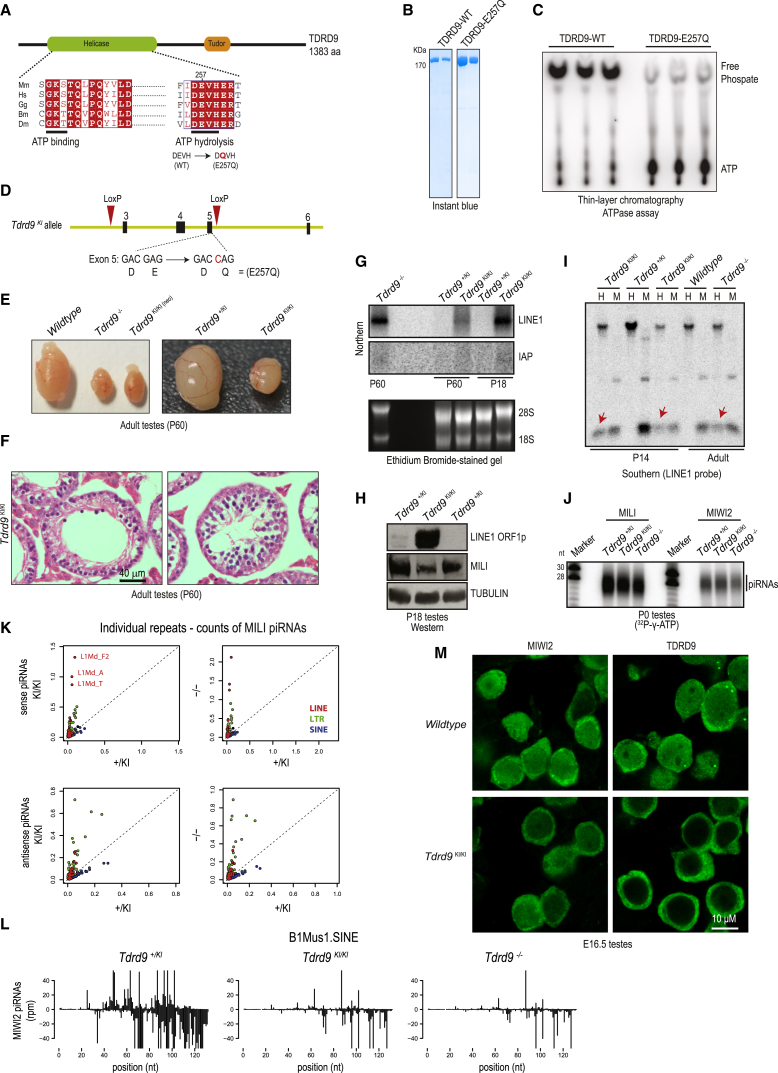
Figure 5.
Mouse TDRD9 Is an ATPase, and Its Activity Is Essential for Transposon Silencing, but Not for piRNA Biogenesis
(A) Domain architecture of mouse TDRD9 with putative consensus amino acid residues responsible for ATP binding and ATP hydrolysis is shown. The point mutation E257Q that abolishes ATPase activity is indicated.
(B) Quality of recombinant mouse TDRD9 protein used for ATPase assays. Wild-type and E257Q point mutant versions were produced.
(C) Thin-layer chromatography of ATPase reactions revealing the faster-migrating free phosphate in the presence of the wild-type TDRD9 protein.
(D) Creation of the catalytic-dead Tdrd9 knockin (KI) mouse carrying the E257Q mutation in the ATPase motif (DEVH → DQVH). The same mouse line also allows creation of the knockout (−/−) mutant, by using loxP sites flanking exons 3–5. See also Figure S6.
(E) Representative image of adult testes from indicated genotypes, showing atrophied testes in homozygous Tdrd9 knockout and knockin mutants.
(F) H&E staining of adult testes from homozygous Tdrd9 knockin mutant showing arrested germ cell development. Scale bar, 40 μm. See also Figure S7A.
(G) Northern analysis of transposons in total testicular RNA, showing derepression of LINE1 retrotransposons in homozygous Tdrd9 knockout and knockin mutants. Age of donor animals is indicated.
(H) Western analysis of total testicular lysates for L1ORF1p expression. MILI (germ cell marker) and TUBULIN (loading control) expression was also examined.
(I) Methylation-sensitive Southern blotting for LINE1 genomic loci. The red arrows point to fragments appearing under conditions of reduced DNA methylation in the homozygous Tdrd9 mutants. H, HpaII-digested DNA; M, MspI-digested DNA.
(J) Immunoprecipitation of PIWI proteins and 5′ end labeling of associated small RNAs from neonatal (P0) testes.
(K) Comparison of MILI-associated piRNAs mapping to individual repeats. There is a striking enrichment of the piRNAs produced from LINE and LTR repeats in Tdrd9 mutants (Tdrd9KI/KI and Tdrd9−/−). See also Figure S8.
(L) Graphs show the distribution of MIWI2-associated piRNAs mapped along B1Mus1.SINE consensus sequence, revealing a depletion of piRNAs in the Tdrd9KI/KI and Tdrd9−/− mutants.
(M) Immunofluorescence analysis of indicated proteins in embryonic testes (embryonic day 16.5) of the different genotypes. Note the nucleo-cytoplasmic distribution of TDRD9 in wild-type germ cells, while it is restricted to the cytoplasm in the Tdrd9KI/KI mutant.