Figure 4.
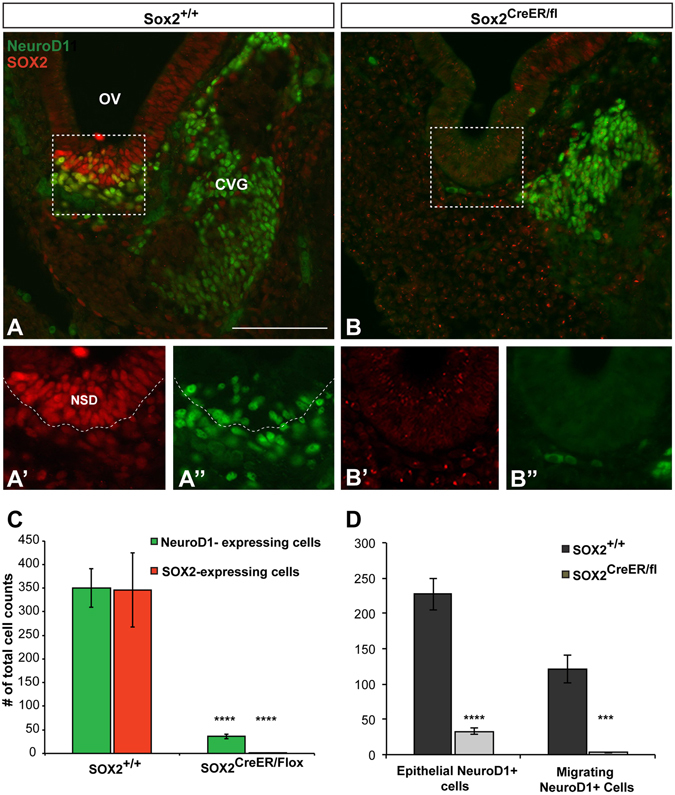
SOX2 is required for the downstream expression of NeuroD1. (A) Sox2 +/+ control section showing the antereoventral portion of the otic vesicle from which neuroblasts delaminate. Boxed region refers to the area in which cell counts were performed. (A’ and A”) show a higher magnification of the neuroblasts contained within the boxed region. Note the extensive colabeling of SOX2 (red) and NEUROD1 (green). Dotted line shows the boundary between the neurosensory epithelium and the mesenchyme. (B) Representative section showing the absence of SOX2 and NEUROD1 protein expression in a Sox2-deficient mutant. (B’ and B”) Show a higher magnification of the boxed region from (B). Note the absence of immunofluorescence for both SOX2 and NEUROD1. To note, the handful of green cells seen in the mesenchyme did not meet inclusion criteria for neuroblasts and were not counted. (C) The total number of NEUROD1 and SOX2-positive neuroblasts was quantified, and both markers were found to be significantly decreased in Sox2 CreER/fl mice (n = 4) compared to Sox2 +/+ controls (n = 3) (****p < 0.0001) (Student’s t test). (D) Total NEUROD1 cell counts were separated based on either being located in the neurosensory epithelium, or migrating into the mesenchyme; in both cases the number of NEUROD1-positive cells were significantly reduced in the Sox2 CreER/fl mice (epithelial NEUROD1: ****p < 0.0001 migrating NEUROD1: ***p = 0.0002). Error bars represent SEM. OV: Otic Vesicle; CVG: Cochleovestibular ganglion; FG: Facial ganglion. Scale bars: 100 μm.
