Figure 4. MCM8 interacts with cyclin D1.
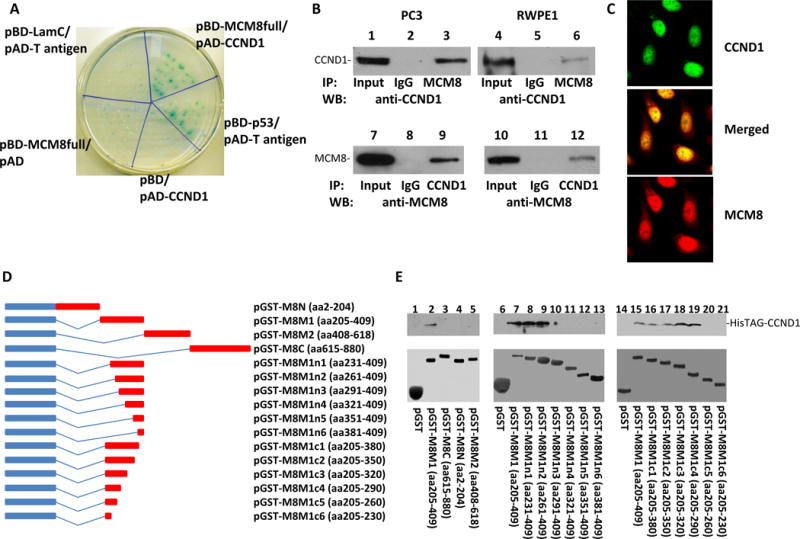
(A) Yeast Two-hybrid screening identified that MCM8 bound cyclin D1. Co-transformants of pBD-MCM8full with pAD-CCND1 on SD agar plate with high stringent nutrient selection (SD-leu-Trp-His-Ade) are shown. Co-transfection of pBD-p53 and pAD-T-antigen is the positive control, while co-transfection of pBD-LamC and pAD-T-antigen, pBD-MCM8full, and pAD, pBD and pAD-CCND1 are the negative controls. (B) Co-immunoprecipitations of MCM8 (lanes 3 and 6) or cyclin D1 (lanes 9 and 12) using antibodies specific for MCM8 or cyclin D1 were performed on protein extracts from PC3 (left panels) and RWPE1 (right panels). The immunoprecipitates were blotted with the indicated antibodies. (C) Immunofluorescence staining of RWPE-1 cells with an antibody against cyclin D1 recognized by FITC- conjugated secondary antibodies for mouse and antibodies specifically against MCM8 recognized by TRITC conjugated secondary antibodies for rabbit. (D) Mapping binding motif of cyclin D1 on MCM8. Left: Schematic diagram of fusion proteins of a series of MCM8 deletion mutants with glutathione-S-transferase (GST) protein. Right: Binding assays of GST or GST-MCM8 deletion mutants with HisTAG-cyclin D1 purified from E.coli. The bound cyclin D1 was blotted with anti-cyclin D1 antibodies. Top panel: Immunoblots with antibodies specific for cyclin D1; Bottom panel: Coomassie staining of the fusion proteins.
