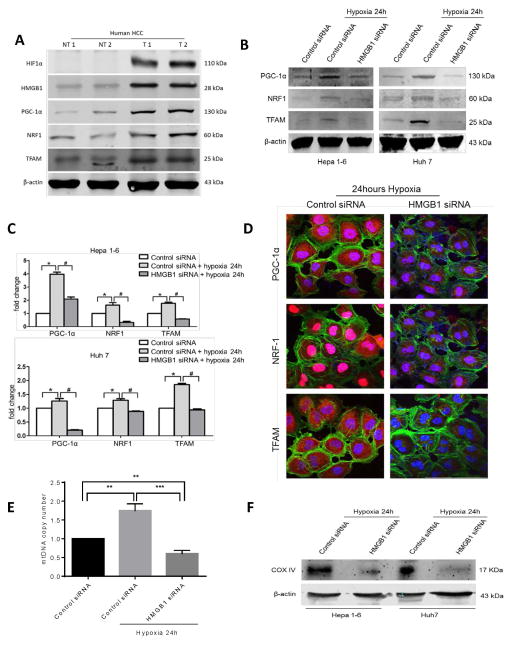
Figure 2. HMGB1 facilitates PGC1-α expression and mitochondrial biogenesis in hypoxic cancer cells.
(A) HIF-1α, HMGB1, PGC-1α, NRF1 and TFAM levels were measured using Western blot analysis in paired human HCC samples and their non-tumor counterparts. In humans there is increased HMGB1 expression and upregulation of mitochondrial biogenesis associated proteins within the hypoxic tumors (NT, nontumor liver; T, Tumor). Selected samples are representative of 15 unique paired samples. (B) There was a significant decrease in the expression of PGC-1α, NRF1 and TFAM when HMGB1 was silenced in the HCC cell lines compared to a scramble control siRNA when cells were subjected to 24h of hypoxia. (C) Real-time PCR analyses of the relative expression of the mitochondrial biogenesis associated genes in hypoxic control HCC cells normalized to normoxic control HCC cells, and hypoxic HMGB1 siRNA HCC cells. *, #p<0.05. (D) Using confocal microscopy, there is a significant decrease in the expression of the mitochondrial biogenesis associated protein in HMGB1 silenced Huh7 cells compared to control siRNA Huh7 cells after 24 hours of hypoxia. Mean PGC1-α area (μm2/nucleus): 16.7 in control versus 4.5 in HMGB1 siRNA; Mean NRF1 area (μm2/nucleus): 1.03×103 in control versus 0.16×103 in HMGB1 siRNA; Mean TFAM area (μm2/nucleus): 66.1 in control versus 12.8 in HMGB1 siRNA; p<0.05. PGC-1α, NRF1, TFAM (red), Nuclei (blue), β-actin (green); Scale Bar: 100μm. (E) Mitochondrial DNA (mtDNA) content in Hep1-6 cells undergoing 24h of hypoxia treated with either HMGB1 siRNA or control siRNA compared with control cells under normoxic conditions (arbitrarily set to 1). **P<0.01, ***P<0.001. (F) Mitochondrial density was also measured by Western blot analysis of mitochondrial protein COX IV in Hep1-6 and Huh7 cells treated with either control or HMGB1 siRNA.