Figure 1.
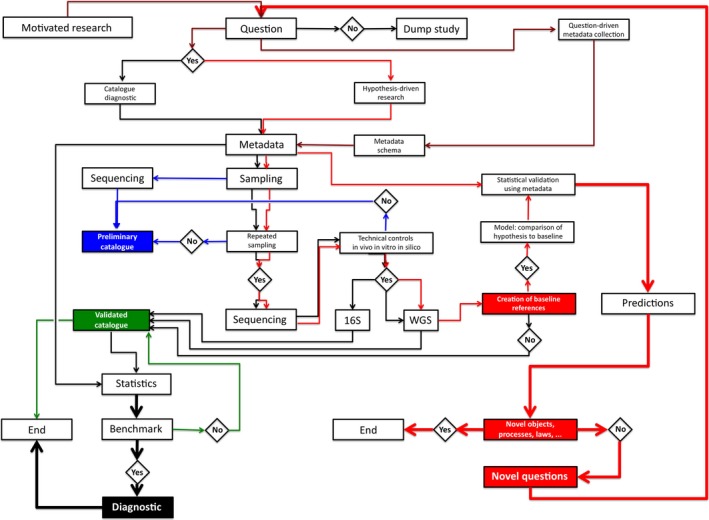
Flow chart of microbiota metagenome studies. Data‐driven research is often purely descriptive. It must start with a specific and clearly stated question, often driven by motivated research (Danchin, 2010). With relevant controls, it may lead to a preliminary catalogue that should be taken with a grain of salt (blue), a validated catalogue (green) or a diagnostic tool (black). Hypothesis‐driven research will require repeated experiments, control experiments, baseline references (the subject of the present article) and model‐building. This will lead investigators to make predictions. If they predict a novel object or process the question can be considered as answered. If not the prediction generates novel questions and the exploration is recursively started again, in a constructive development, typical of what science should be. An essential part of the process is the creation and management of metadata that are linked to the sampling procedure. Connection with the metadata allows investigators to place their questions and answers in relevant context. Sequencing is key to microbiome studies. Here, we considered that restricting sequencing to 16S targets will restrict the study to catalogues and diagnostic. This approach can seldom be used in hypothesis‐driven research, where whole‐genome sequencing (WGS) must be the rule.
