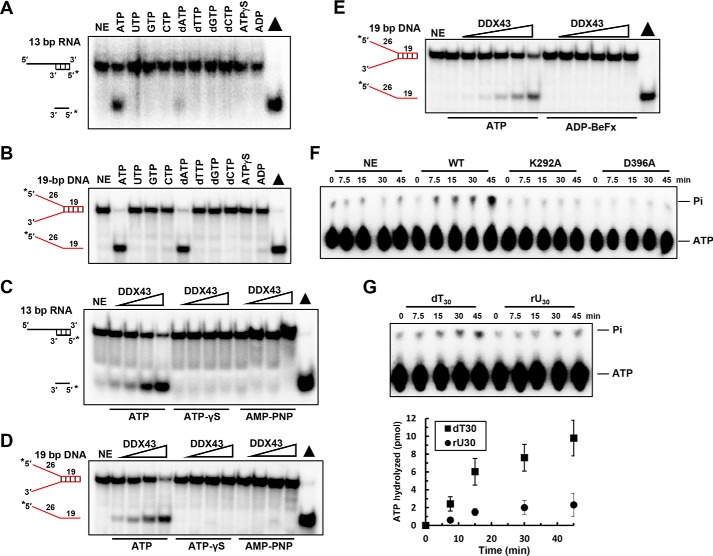
Figure 4.
ATP hydrolysis is required for DDX43 unwinding. A and B, representative images of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate (A) or forked 19-bp duplex DNA substrate (B) with 150 nm DDX43 protein at 37 °C for 15 min with different nucleoside triphosphates. NE, no enzyme. C and D, representative images of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate (C) or forked 19-bp duplex DNA substrate (D) with ATP or ATP analogs with increasing protein concentrations (0–3 μm). E, representative images of helicase reactions performed by incubating 0.5 nm 19 bp forked dsDNA substrate with increasing protein concentrations of DDX43 (0–3 μm) and 2 mm ATP or ADP-BeFx at 37 °C for 15 min. F, a representative image of ATP hydrolysis detected by TLC with DDX43 protein (2.5 μm) with M13 ssDNA effector (50 μm). WT, wild type; K292A and D396A are two engineered mutants. G, a representative image (top) of DDX43 protein ATP hydrolysis detected by TLC with indicated nucleic acid cofactor dT30 or rU30 (30 μm) and quantitative analyses of ATP hydrolysis (bottom). Data represent the mean of at least three independent experiments with S.D. indicated by error bars.