Figure 2.
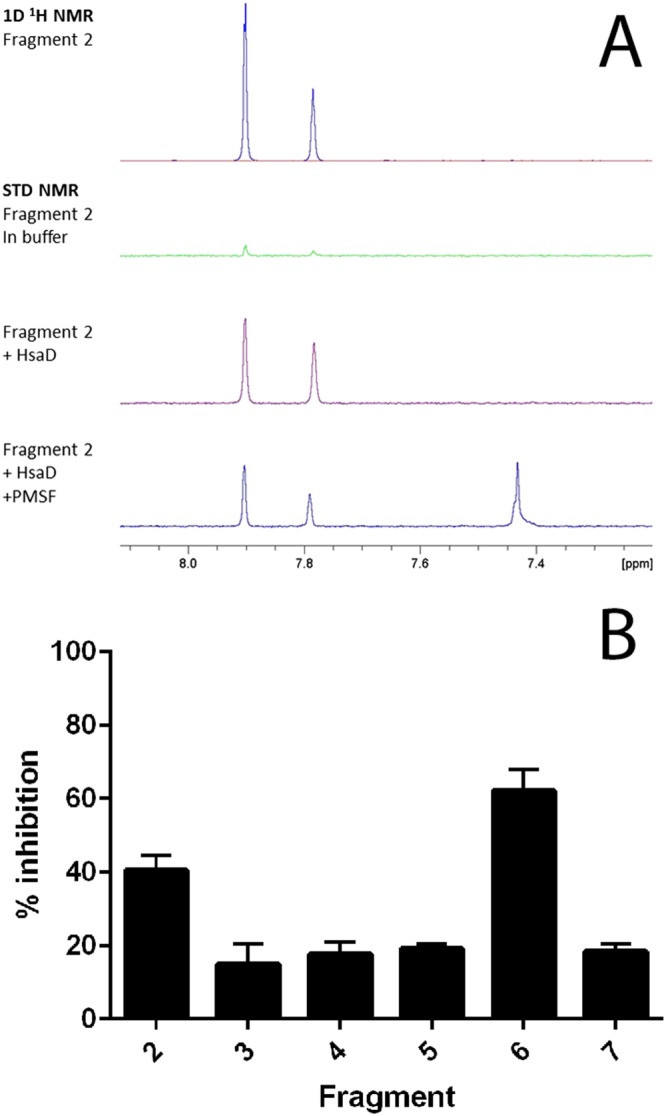
(A) NMR evidence of fragment binding. Water‐suppressed 1D 1H and STN NMR screening spectra for a representative fragment hit (fragment 2, at 1.2 mM) in the presence and absence of HsaD (10 μM) plus PMSF (100 μM). Only the fragment resonances in the aromatic region of the spectra are shown. The strong positive fragment signals in the presence of HsaD (purple spectrum) indicate binding, and this interaction is reduced by the addition of PMSF. (B) Inhibition of HsaD by DSF hits. Fragments identified by DSF as binding to HsaD were assayed for inhibition as described in Methods at 1 mM. The mean of six independent experiments (each carried out in triplicate) ± SD is shown. Fragments are numbered as shown in Table 1; 100% activity was defined as the rate of enzymatic HOPDA hydrolysis by 16 μg·mL−1 HsaD with vehicle (no compound), which was 0.08 μmol·min−1·(mg protein)−1.
