Table 1.
Fragments from a 1258 member library producing a significant shift in HsaD TM and where saturation NMR demonstrated displacement of the ligand by PMSF
| Compound | TM shiftsa | NMR | PMSF displacement | Structure |
|---|---|---|---|---|
| 1 | 1.78; 2.64; 2.64 | Hit | Displaced |
(1).
|
| 2 | 0.77; 0.38; 0.38 | Hit | Displaced |
(2).
|
| 3 | 0.55; 0.76; 0.76 | Hit | Displaced |
(3).
|
| 4 | 0.55; ‐2.26; ‐2.26 | Hit | Displaced |
(4).
|
| 5 | 0.92; 1.13; 1.13 | Hit | Not displaced (PMSF bound) |
(5).
|
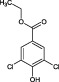
| 6 | 0.70; 1.13; 1.13 | Hit | Not displaced (PMSF not bound) |
(6).
|
| 7 | 0.82; 0.38; 0.76 | Hit | Displaced |
(7).
|
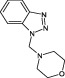
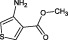
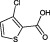
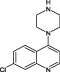
The compounds are numbered sequentially: 4‐((1H–benzo[d][1,2,3] triazol‐1‐yl)methyl)morpholine compound 1; 3,5,‐dichloro‐benzenesulfonamide compound 2; methyl 4‐aminothiophene‐3‐carboxylate compound 3; 3‐chlorothiophene‐2‐carboxylic acid, compound 4; 7‐chloro‐4‐(piperazin‐1‐yl)quinolone, compound 5; ethyl 3,5‐dichloro‐4‐hydroxybenzoate, compound 6; 4‐(piperidin‐1‐ylsulfonyl)aniline, compound 7.
The TM shifts in °C are shown for each compound tested with a positive result for DSF, performed as described in the Methods section at 2.5 mM final fragment concentration per well. Structures are drawn with ChemDraw Ultra 12.0 (CambridgeSoft). Hits from the primary DSF screen were re‐tested twice to confirm results of the initial screen and finally validated for protein binding using NMR spectroscopy (as shown in Figure 2A) (after Ciulli and Abell, 2007; Mayer and Meyer, 1999).