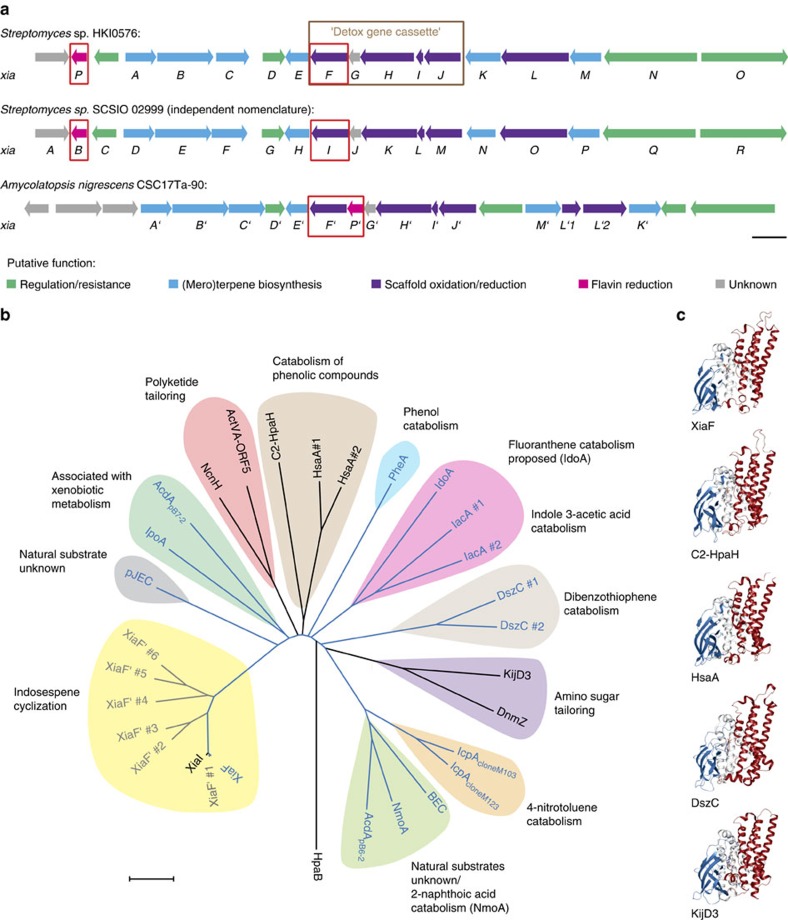
Figure 2. Comparison of xia gene loci, and phylogeny of XiaF and its homologues.
(a) Comparison of the xia gene cluster of Streptomyces sp. (HKI0576) with the xia gene cluster of Streptomyces sp. SCSIO 02999 and a putative xia gene locus of Amycolatopsis nigrescens CSC17Ta-90. Genes for the two-component system XiaF/XiaP and orthologues are framed in red. The ‘detox gene cassette’ xiaFGHIJ, which encodes homologues of xenobiotic-metabolizing enzymes that are mostly involved in IST tailoring reactions is framed in brown. The length of the scale bar represents a kilobase. (b) Phylogenetic tree of XiaF and related flavoenzymes. Enzymes that have been reported to catalyse indigo formation are coloured in blue; putative XiaF orthologues of other putative xia gene clusters that yet have not been characterized are coloured in grey. The evolutionary history was inferred using the neighbour-joining method36. The oxygenase component HpaB of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8 (ref. 90) was used as an outgroup. The length of the scale bar represents an evolutionary distance of 0.1 amino acid differences per site. (For further details, see the Supplementary Note 3 and Supplementary Figs 8–11.) (c) Structural comparison of XiaF (PDB ID: 5LVW), the 4-hydroxyphenylacetate 3-hydroxylase C2-HpaH (PDB ID: 2JBT)48, the secosteroid hydroxylase HsaA from Mycobacterium tuberculosis (PDB ID: 3AFE)32, the dibenzothiopene monooxygenase DszC from Rhodococcus erythropolis D-1 (PDB ID: 3X0Y)52 and the N-oxygenases KijD3 involved in D-kijanose biosynthesis (PDB ID: 4KCF)53. The structural alignment was calculated by the Dali server51 and shows the superposition of the different monomers. (For a more detailed structural comparison, see Supplementary Fig. 7.)