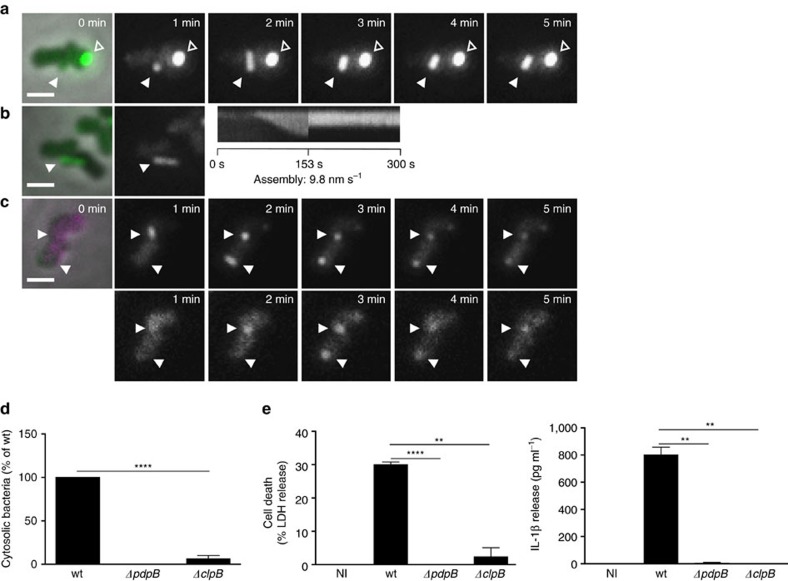
Figure 3. Phagosomal rupture and AIM2 inflammasome activation is dependent on disassembly of T6SS sheaths by ClpB.
(a) T6SS dynamics in F. novicida U112 iglA-sfGFP ΔclpB. Arrowheads indicate T6SS sheath assembly, contraction and location of sheath after contraction. Empty arrowheads indicate non-dynamic IglA-sfGFP foci. First image is a merge of phase contrast and GFP channels, following images represent GFP channel only. (b) Kymogram of F. novicida U112 iglA-sfGFP ΔclpB over 5 min (3 s per pixel). First image is a merge of phase contrast and GFP channels, following images represent GFP channel only. (c) Colocalization of ClpB-mCherry2 with IglA-sfGFP (arrows) in F. novicida U112 iglA-sfGFP clpB-mCherry2. First image is a merge of phase contrast, GFP and mCherry channels, following images represent GFP channel (upper panel) and mCherry channel (lower panel). (d) Quantification of cytosolic bacteria in unprimed wild-type BMDMs 4 h after infection with F. novicida U112 iglA-sfGFP wild type, ΔpdpB or ΔclpB (normalized to wild type). (e) Release of LDH and mature IL-1β from primed wild-type BMDMs 10 h after infection with F. novicida U112 iglA-sfGFP wild type, ΔpdpB or ΔclpB (NI—noninfected control). (a–c) 3.3 × 3.3 μm fields of view are shown. Scale bars, 1 μm. (d,e) Data are pooled from three independent experiments (d) (mean and s.d. are shown) or representatives of three independent experiments (e) (mean and s.d. of triplicate wells are shown). **P<0.01 and ****P<0.0001 (two-tailed unpaired t-test with Welch’s correction).