Fig. 1.
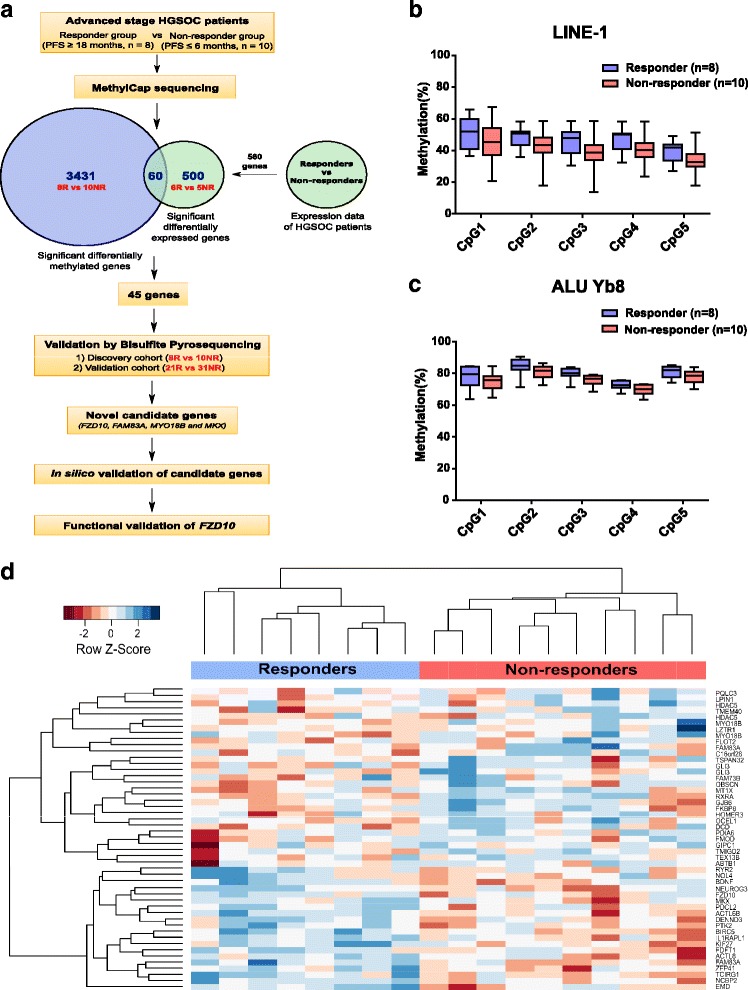
Identification of novel DNA methylation genes by using MethylCap-seq between extreme responder and non-responder HGSOC patients. a Experimental strategy to evaluate differential DNA methylation regions (DMRs) between extreme chemoresponse patient groups and their subsequent validation. b and c Bisulfite pyrosequencing for global methylation marker LINE-1 and ALU Yb6 in responder and non-responder groups showing similar global methylation level. Each bar represents average methylation in % ± SD of either responder (n = 8) or non-responder (n = 10) at a specific CpG site. d Hierarchical clustering of significant DMRs (49) in the responders (n = 8) and non-responders (n = 10) in the discovery set (Set 1)
