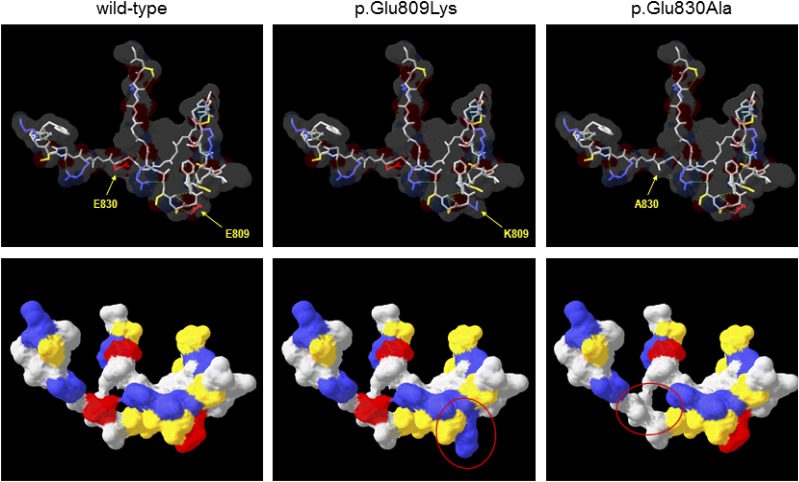
Figure 2.
Comparative protein modeling of WFS1. The COOH-terminal lumenal domain of WFS1 (residues 653–869) was submitted to SWISS-MODEL for template identification; models were then constructed for residues 798–838 using the top-scoring template (PDB identifier 1ltl; MCM protein from Methanothermobacter thermautotrophicus; 27% sequence similarity over region modeled). Upper panels show the protein backbone with selected side chains as indicated in stick format for wild-type and variant WFS1, the predicted molecular surface is shown as a transparent layer, and broken green lines indicate predicted hydrogen bonds. Lower panels show the same structures but rotated ∼45° away from the viewer around a horizontal axis and with the predicted molecular surface shown as a solid layer. In all panels, amino acids are colored by type (red, acidic; blue, basic; yellow, uncharged polar; gray, nonpolar/hydrophobic); regions of significantly altered surface properties are indicated by red ovals in the lower panels. A830, p.Ala830; E809, p.Glu809; E830, p.Glu830; K809, p.Lys809.