Figure 2. Flow chart illustrating sample preparation and analysis by shotgun and targeted mass spectrometry.
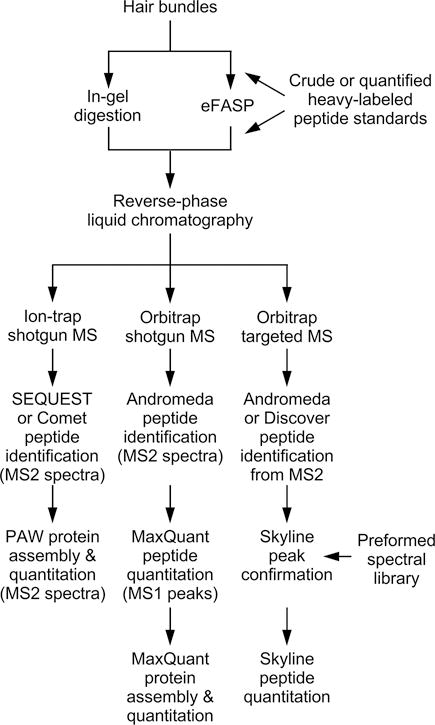
Hair bundle proteins can be prepared for mass spectrometry using either in-gel or eFASP digestion; if used, standards (e.g., heavy-isotope labeled peptides) are added after digestion. Peptides are separated by reverse-phase liquid chromatography, introduced into the mass spectrometer by electrospray ionization, and are subjected to tandem mass spectrometry. If data are acquired on an ion-trap mass spectrometer, MS2 spectra are analyzed by Comet peptide searching and PAW protein quantitation. Shotgun data acquired on an Orbitrap (high resolution) mass spectrometer are analyzed by Andromeda peptide searching of MS2 data and MaxQuant protein quantitation using MS1 peak intensities, while targeted data are analyzed by Skyline after Andromeda or Protein Discoverer peptide searching.
