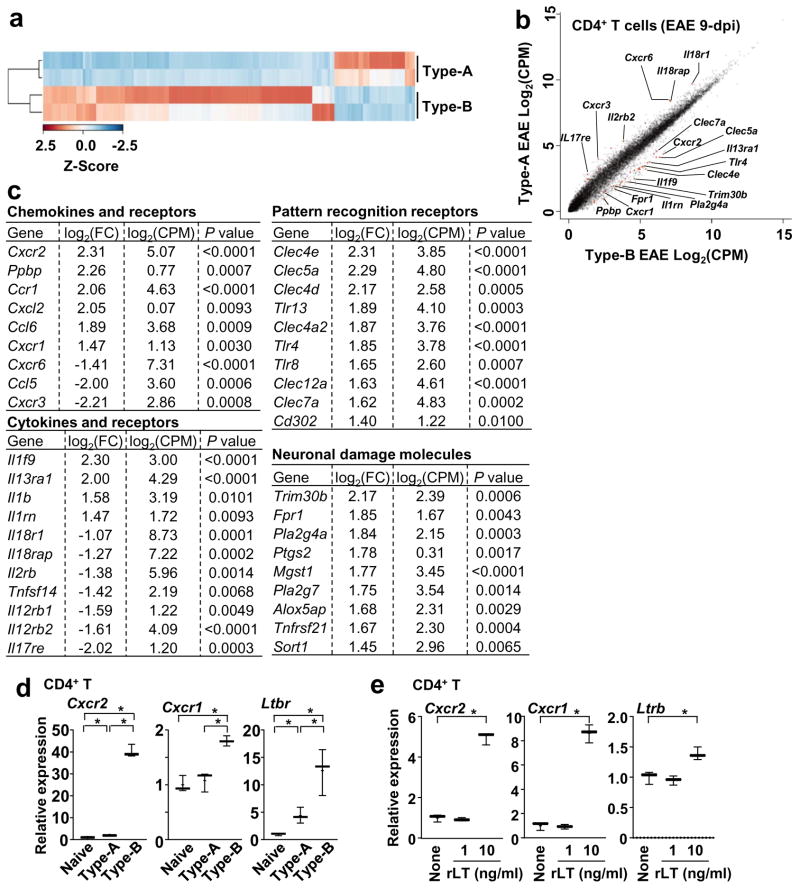
Figure 3. Gene expression profile of CD4+ T cells between Type-A and Type-B EAE.
CD4+ T cells were FACS-purified from spleens at 9-dpi and submitted for the RNA-seq analysis. (a) Heatmap of of 1,846 differentially expressed genes (P<0.05; log2FC >1 or <−1) between CD4+ T cells from Type-A and Type-B EAE mice. (b) Scatter plot comparing transcriptome. (c) Selected genes from pools with P<0.01, log2FC >1 or <−1, and a log2CPM>1. (d) mRNA levels of Cxcr2, Cxcr1 and Ltbr in splenic CD4+ T cells obtained from naïve mice and mice with Type-A or Type-B EAE induction at 9-dpi determined by qPCR. (n=3). Cxcr2: P(Naive vs Type-A)=0.0231, t(4)=3.583, P(Naive vs Type-B) <0.0001, t(4)=23.22, P(Type-A vs Type-B) <0.0001, t(4)=22.75. Cxcr1: P(Naive vs Type-A)=0.5999, t(4)=0.5689, P(Naive vs Type-B)=0.0015, t(4)=7.806, P(Type-A vs Type-B)=0.0036, t(4)=6.119. Ltbr: P(Naive vs Type-A)=0.0170, t(4)=3.937, P(Naive vs Type-B)=0.0091, t(4)=4.733, P(Type-A vs Type-B)=0.0333, t(4)=3.188. (e) Levels of Cxcr2, Cxcr1 and Ltbr mRNA in CD4+ T 6h after rLT1 treatment at indicated concentrations. (n=3). P(Cxcr2) <0.0001, t(4)=19.36, P(Cxcr1) <0.0001, t(4)=16.09, P(Ltbr)=0.0116, t(4)=4.413. All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. Data sets (d, e) are representatives from at least 2 similar experiments for each.