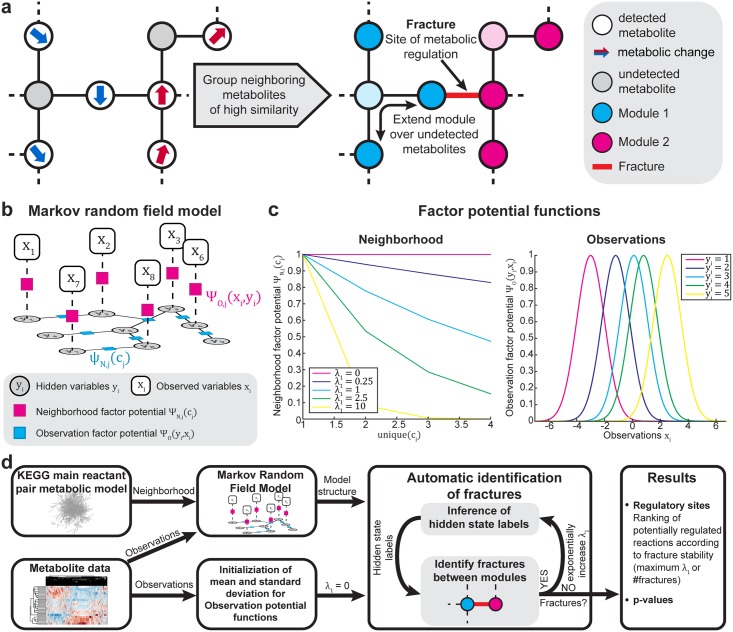
Fig 1. Design and implementation of the metabolic network segmentation algorithm, a probabilistic graphical modelling approach to identify sites of metabolic regulation.
(a) Principle of the algorithm to identify sites of metabolic regulation using metabolomics data and metabolic network reconstructions. The algorithm segments the metabolic network into modules of highly similar changes. To deal with sparse and noisy metabolite measurements, it considers dependencies between neighboring metabolites to extend the modules. Fractures between two modules are potential sites of metabolic regulation. (b) Structure of the Markov random field model. The discrete hidden variables y represent the modules to which a metabolite belongs, the observed variables x integrate the metabolite measurements. The factor potential functions introduce probabilistic dependencies between neighboring hidden states (ψN) and between measurements and the hidden states (ψO). (c) Example of neighborhood and observation factor potential functions. The neighborhood factor potentials are module label dependent exponential decay functions. The influence of the metabolic neighborhood is controlled by λ1. Observation potential functions link hidden state/module labels to metabolic observations by hidden state dependent Gaussian distributions. (d) Algorithm workflow of the scan mode implementation to automatically identify sites of metabolic regulation. Details in S1 Fig.