Figure 1.
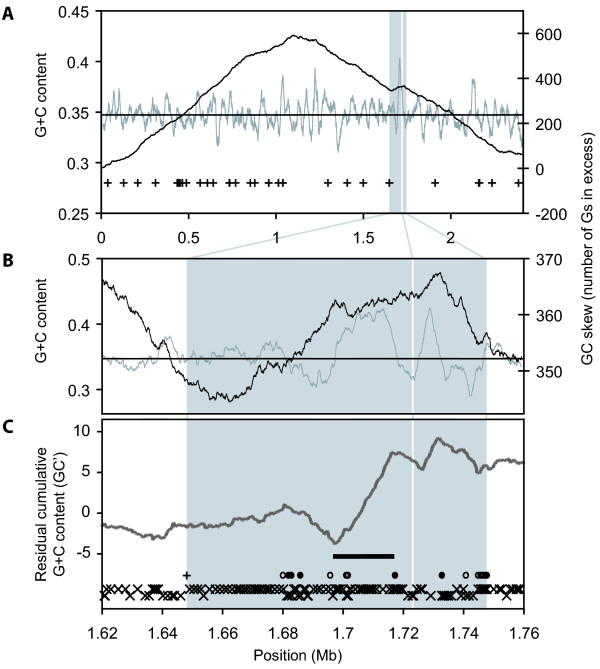
The genomic island (GI) present in the chromosome of the endosymbiont UWE25. (A) Position of the GI on the UWE25 genome, a 100-kb region (grey area) delimited by two direct repeats (Table 1) at both ends and by two gly-tRNAs genes in tandem (all tRNAs genes are represented by '+') at its proximal end. A third copy of the direct repeat (Table 1) is indicated by a white line disrupting the grey area. The region is characterized by a different slope in the cumulative GC skew analysis (black curve) and by a higher G+C content (grey curve, windows of 20 kb, 0.1-kb step). The horizontal line indicates the genomic G+C content average. (B) Closer view of the 100-kb region (black curve, cumulative GC skew; grey curve, G+C content windows of 5 kb, 0.1-kb step; horizontal line, average genomic G+C content). (C) Residual cumulative G+C content (GC') and genomic features of the 100-kb GI. This region encompasses the region with the highest G+C content in the 20-kb windows analysis of the UWE25 genome. The position of genes is represented by an 'X' on the upper line if encoded on the positive strand, otherwise by an 'X' on the bottom line (For details, see Table in Supplementary Material 1). A large majority of genes are co-oriented in the genome region flanked by the direct repeats. The tra operon (thick line), present on this GI, exhibits a G+C content (40.0%) clearly higher than that of the whole genome (34.7%). The positions of transposases (open circles) and of phage-related genes (full circles) are indicated.
