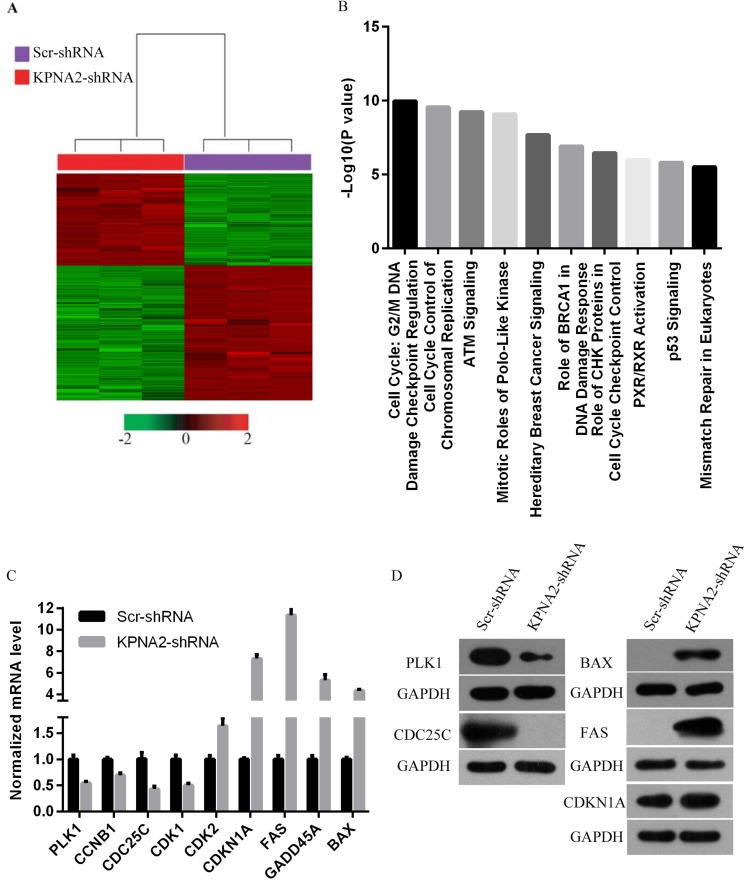
Figure 7. Widespread changes in gene expression and pathways essential for tumorigenesis in human hepatocellular carcinoma cell line HepG2 with KPNA2 knockdown revealed by microarray analysis.
(A) Heat map representation of 1585 genes with significant differential expressions in HepG2 human hepatocellular carcinoma cells infected with lentiviruses expressing either Scr-shRNA (purple) or KPNA2-shRNA (red) under the significance criteria of P < 0.05 and | fold change | > 1.5. Genes and samples are listed in rows and columns, respectively. A color scale for the normalized expression data are shown at the bottom of the microarray heat map (green represents downregulated genes, while red represents upregulated genes). (B) Functional pathway enrichment was analyzed using IPA. Here, the top 10 enriched pathways were shown. The statistical significance shown on the x-axis is represented by the inverse log of the P value. (C) Confirmation of microarray data using real-time quantitative PCR for selected genes PLK1, CCNB1, CDC25C, CDK1, CDK2, CDKN1A, FAS, GADD45A and BAX in HepG2 human hepatocellular carcinoma cells infected with lentiviruses expressing either Scr-shRNA or KPNA2-shRNA. The data shown here are from one of three independent experiments (p < 0.01) and are normalized to GAPDH. (D) Protein level of PLK1, CCNB1, CDC25C, CDK1, CDK2, CDKN1A, FAS, GADD45A and BAX in HepG2 human hepatocellular carcinoma cells infected with lentiviruses expressing either Scr-shRNA or KPNA2-shRNA. GAPDH was used as an internal control.