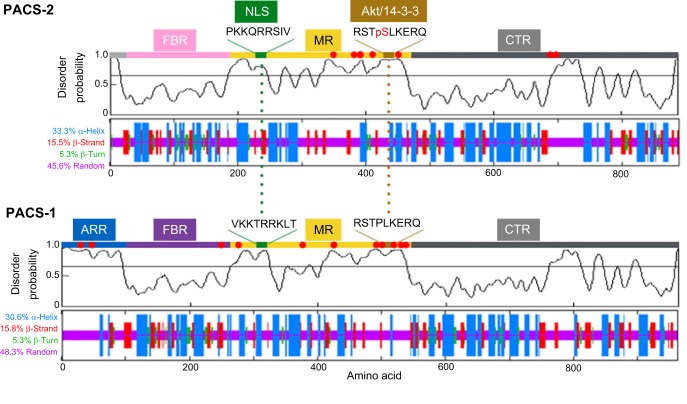
Fig. 1.
Disorder prediction for PACS-1 and PACS-2. The PrDOS server (http://prdos.hgc.jp/cgi-bin/top.cgi; Ishida and Kinoshita, 2007) was used to predict natively disordered regions from the amino acid sequences of the human PACS-1 (UniProt Q6VY07) and PACS-2 (UniProt Q86VP3) proteins [false discovery rate (FDR)=2%]. The disorder probabilities for each residue were plotted as a function of length and the graphical profiles were juxtaposed with the predicted secondary structures, which were obtained using an improved self-optimized prediction method (SOPMA) on a set of aligned members of the PACS-1 or PACS-2 protein families (lower plots). The PACS-2 nuclear localization signal (NLS) and Akt site, which binds 14-3-3 proteins, together with the corresponding sequences in PACS-1 are shown and predicted to reside in disordered regions. ARR, atrophin-1-related region; FBR, furin (cargo)-binding region; MR, middle region; CTR, C-terminal region. Red dots, phosphorylation sites [as predicted by PhosphoSitePlus (http://www.phosphosite.org/; Hornbeck et al., 2015)].