Fig. 2.
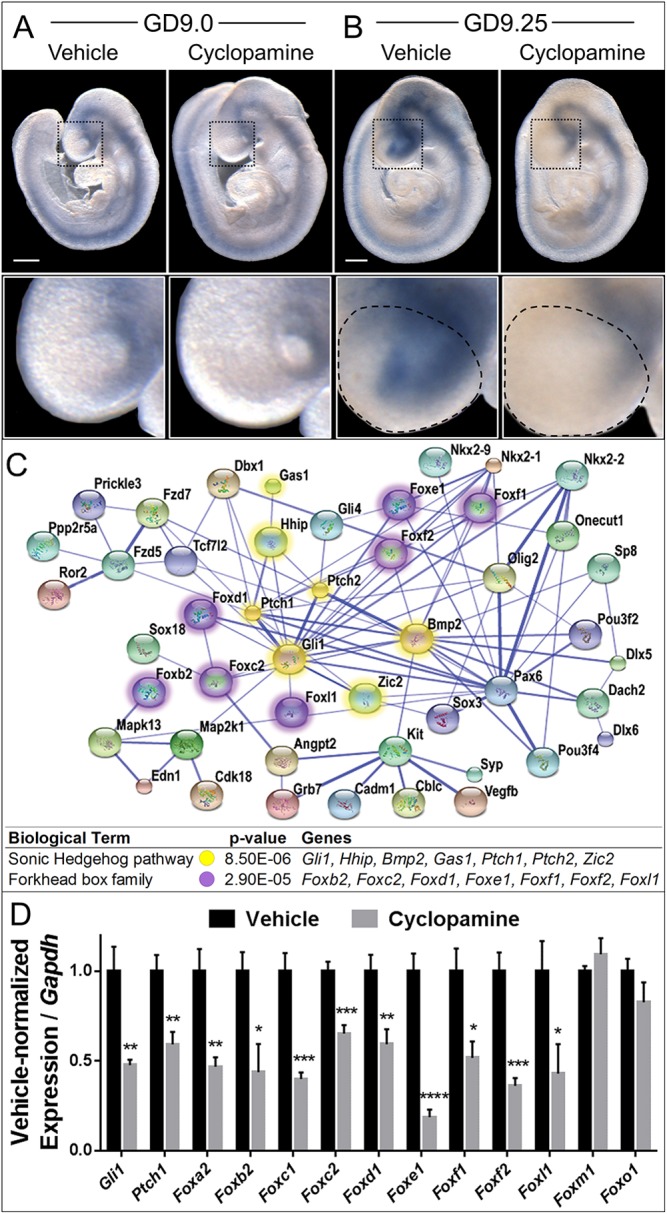
Shh regulates Fox genes in cleft lip pathogenesis. (A,B) Stage-matched GD9.0 (A) and GD9.25 (B) embryos exposed in utero to vehicle or cyclopamine and subjected to ISH for Gli1. The boxed region of the FNP is magnified beneath. Although Gli1 expression is grossly similar at GD9.0, downregulation of Gli1 is apparent in cyclopamine-exposed embryos by GD9.25. The microdissected FNP region used for the microarray and subsequent RT-PCR validation is outlined on the GD9.25 embryos. Scale bars: 300 μm. (C) A STRING network of select significantly differentially expressed genes identified by microarray. Colored nodes represent significant dysregulated genes; blue edges show functional and predicted interactions between gene products, with the thicker, darker lines representing interactions for which evidence is more abundant. Note the high degree of centrality of Shh signaling pathway genes (yellow) and Forkhead box (Fox) transcription factor genes (purple). P-value indicates significant enrichment for the Shh pathway and Fox family within the dysregulated gene set. (D) RT-PCR validation of the indicated genes in vehicle-exposed or cyclopamine-exposed FNP tissue. Values represent mean±s.e.m. expression of the cyclopamine-exposed group relative to Gapdh, normalized to the vehicle control group (n=6 pooled litters per group). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (two-tailed t-test with Holm-Sidak correction).
