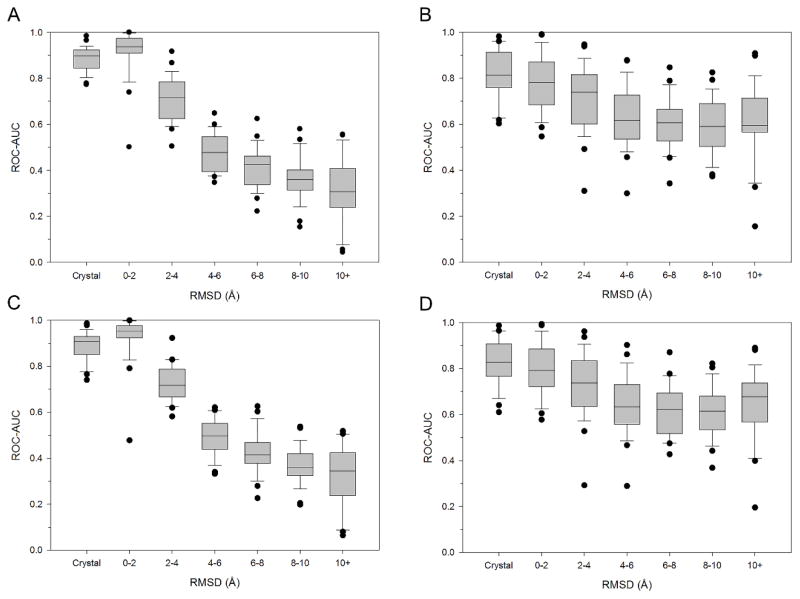
Figure 1. Enrichment power versus binding pose accuracy across 26 kinase targets.
A set of 20 unique binding poses was generated for 940 co-crystallized inhibitors across 26 kinase targets in PDBBind’s general set. The RMSD to the native crystal pose was used to separate the binding poses into the following bins: <2, 2–4, 4–6, 6–8, and >10 Å. ROC-AUC performance are shown in a box-and-whisker plot for each of the docking method and scoring function combinations: (A) Glide/GlideScore; (B) Glide/SVMGen; (C) Vina/GlideScore; (D) Vina/SVMGen.