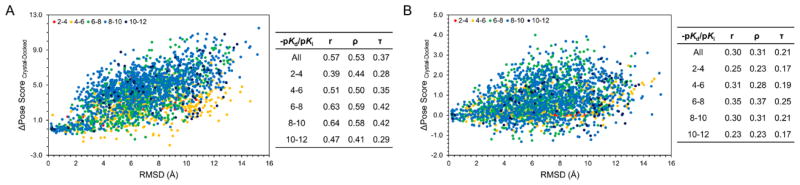
Figure 2. Binding pose accuracy versus deviation in predicted scores.
A set of 20 unique binding poses was generated for 123 co-crystallized kinase inhibitors from PDBBind’s refined set. For each pose, the difference between the crystal and docked scores was plotted against the RMSD between the docked and crystal poses. Compounds were binned based on their experimental binding affinities into ranges of 2–4, 4–6, 6–8, 8–10, and 10+. Pearson, Spearman, and Kendall correlations were calculated overall and for each bin for (A) GlideScore and (B) SVMGen.