Abstract
BACKGROUND
Diabetes, obesity, and overweight are prevalent pregnancy complications that predispose offspring to neurodevelopmental disorders, including autism, attention-deficit/hyperactivity disorder, and schizophrenia. Although male individuals are three to four times more likely than female individuals to develop these disorders, the mechanisms driving the sex specificity of disease vulnerability remain unclear. Because defective placental insulin receptor (InsR) signaling is a hallmark of pregnancy metabolic dysfunction, we hypothesized that it may be an important contributor and novel mechanistic link to sex-specific neurodevelopmental changes underlying disease risk.
METHODS
We used Cre/loxP transgenic mice to conditionally target InsRs in fetally derived placental trophoblasts. Adult offspring were evaluated for effects of placental trophoblast-specific InsR deficiency on stress sensitivity, cognitive function, sensorimotor gating, and prefrontal cortical transcriptional reprogramming. To evaluate molecular mechanisms driving sex-specific outcomes, we assessed genome-wide expression profiles in the placenta and fetal brain.
RESULTS
Male, but not female, mice with placental trophoblast-specific InsR deficiency showed a significantly increased hypothalamic-pituitary-adrenal axis stress response and impaired sensorimotor gating, phenotypic effects that were associated with dysregulated nucleotide metabolic processes in the male prefrontal cortex. Within the placenta, InsR deficiency elicited changes in gene expression, predominantly in male mice, reflecting potential shifts in vasculature, amino acid transport, serotonin homeostasis, and mitochondrial function. These placental disruptions were associated with altered gene expression profiles in the male fetal brain and suggested delayed cortical development.
CONCLUSIONS
Together, these data demonstrate the novel role of placental InsRs in sex-specific neurodevelopment and reveal a potential mechanism for neurodevelopmental disorder risk in pregnancies complicated by maternal metabolic disorders, including diabetes and obesity.
Keywords: Epigenetic, Insulin, Prefrontal cortex, Prenatal, Serotonin, Sex
Diabetes, obesity, and overweight during pregnancy are prevalent risk factors for offspring neurodevelopmental disorders, including autism, attention-deficit/hyperactivity disorder (ADHD), and schizophrenia (1–8). Such pregnancy complications confer significant risk to offspring in the United States in particular, where one-third of reproductive-aged women are obese and more than 9% of pregnancies are affected by gestational diabetes (9–11). Male offspring are especially at risk because they are more vulnerable to prenatal insults than female offspring and are three and four times more likely to develop ADHD and autism, respectively (12,13). Animal models have identified fetal sex as a key determinant of lifelong outcomes; however, the molecular mechanisms mediating such sex-specific programming remain unclear (14–18). Impaired insulin action is common to these maternal metabolic conditions, where reduced insulin production is a hallmark of type 1 diabetes mellitus and impaired cellular responses to insulin are characteristic of type 2 diabetes, gestational diabetes, and obesity. Insulin dysfunction has been demonstrated in placental tissue from pregnancies complicated by diabetes, preeclampsia, intrauterine growth restriction, and inflammation (19–27). Critically, then, placental insulin signaling may serve as a novel mediator of neurodevelopmental programming by maternal adversity contributing to disease risk.
Throughout pregnancy, the placenta is important for fetal support because it delivers nutrients and growth factors, maintains a protective barrier, and initiates adaptive responses to intrauterine status signals (28–31). Insulin dynamically regulates placental function across gestation, promoting placental growth, angiogenesis, metabolism, and hormone secretion, especially during early pregnancy (32–34). Perturbation of these processes can elicit distinct responses in male and female individuals that can influence neurodevelopment throughout the entire course of gestation and may therefore underlie the sex-biased outcomes reported in animal studies (35). These unique strategies are likely mediated, in part, by X- and Y-linked gene expression by the fetally derived trophoblasts comprising the majority of the placenta (36,37).
To determine a novel mechanistic link between placental trophoblast-specific insulin receptor (InsR) dysfunction and sex-biased neurodevelopmental programming, we used the Cre/loxP system to conditionally ablate the InsR gene in fetally derived placental trophoblasts (38). Male and female mice were evaluated during adulthood for effects of placental trophoblast-specific InsR deficiency (pKO) on stress sensitivity, cognitive function, sensorimotor gating, and prefrontal cortical reprogramming, critical end points related to neurodevelopmental disorders. We hypothesized that sex differences in placental response to InsR deficiency would promote greater deficits in male individuals than in female individuals. To evaluate potential molecular mechanisms driving sex-specific programming, we assessed genome-wide expression profiles in the placenta and corresponding fetal brain.
METHODS AND MATERIALS
Animals and Conditional InsR Deletion
Placental trophoblast-specific Cre recombinase–expressing mice (CYP19-Cre+; 129S1/SvlmJ background) (38) were bred with floxed InsR mice (Insrtm1Khn/J; C57BL/6J background), and their double heterozygous female (CYP19-Cre+; Insrflox/wt) and Insrflox/wt male offspring were subsequently crossed to generate placental-specific InsR knockout [(pKO) CYP19-Cre+; Insrflox/flox] and control [(pWT) CYP19-Cre−; Insrflox/wt] littermates. Noon on the day of copulation plug detection denoted embryonic day (E) 0.5. Dams were either dissected at E12.5 or E17.5 or allowed to give birth. Mice were weaned into same-sex and same-genotype cages of 2 or 3 on postnatal day (P) 28, weighed every 4 weeks thereafter, and tested beginning at 8 weeks of age. In all experiments, no more than 2 littermates were included in each group (detailed in Supplemental Table S1). Mice were maintained on a 12-hour light/dark cycle (lights on at 07:00) with ad libitum food access (Purina Rodent Chow, Purina Mills, Gray Summit, MO); 28.1% protein, 59.8% carbohydrate, and 12.1% fat) and water. All animal procedures were conducted in agreement with the Guide for the Care and Use of Laboratory Animals in accordance with National Institutes of Health guidelines and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Embryonic Dissection
Timed pregnant dams were deeply anesthetized with isoflurane on E12.5 or E17.5, and conceptuses were isolated from the uterine wall as described previously (39). Intrauterine position, observed resorptions, and tissue wet weights were recorded. Placentas were hemisected in the transverse plane, flash frozen in liquid nitrogen, and stored at −80°C until RNA and protein extraction. Fetal brain, liver, and hindlimb were also isolated, flash frozen, and stored. All dissections were completed between 10:00 and 14:00. Embryonic tissue was retained for determination of sex by Jarid1 genotyping as described previously (39).
Metabolic and Behavioral Phenotyping
One cohort (n = 9–14/group) underwent light–dark exploration, hypothalamic-pituitary-adrenal (HPA) axis assessment, prepulse inhibition (PPI) of acoustic startle, and glucose tolerance test protocols separated by an intertest interval of ≥ 7 days. Brains were harvested from this cohort 2 weeks after the glucose tolerance test. A second cohort (n = 8–10/group) was assessed for spatial learning in the Barnes maze and auditory fear conditioning. Testing was initiated at 8 weeks of age. Protocols are detailed in the Supplement.
Cytochrome C Oxidase Activity
Test-naïve mice (n = 7–8/group) were killed by cervical dislocation at 12 weeks of age, whole brains were collected on dry ice, and prefrontal cortex (PFC) micropunches were isolated using a 1.0-mm Harris Uni-Core tissue puncher (Ted Pella, Redding, CA) as described previously (39) and stored at −80°C until assay. Punches were incubated for 5 minutes in 50 μL of 25 mM potassium phosphate buffer and 2 μL of 10% lauryl maltoside and then centrifuged at 14,000 rpm for 10 minutes at 4°C. Supernatant was moved to a new tube. Then, 10 μL was added to a cuvette containing 0.5 mL of potassium phosphate buffer and 25 μL of reduced cytochrome C. Optical density was measured for 1 minute, and rate of cytochrome C oxidation was calculated using Cary Win kinetics software (Agilent Technologies, Santa Clara, CA) and normalized to total protein determined by Pierce BCA assay (Thermo Fisher Scientific, Waltham, MA).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was reverse transcribed to complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Waltham, MA). Gene expression was determined by TaqMan Gene Expression Assays for the target gene using the 7500 Fast Real-Time PCR System (Applied Biosystems). Raw threshold cycle (Ct) values for each sample were normalized to Actb and analyzed using the comparative Ct method (40).
Western Blot
Following Trizol–chloroform isolation of E12.5 placenta RNA, protein was extracted, quantified, separated by gel electrophoresis, transferred, and probed for InsR as detailed in the Supplement.
Affymetrix Microarray
Total RNA was extracted from placental hemisections and PFC micropunches as described previously (39) and was sent to the University of Pennsylvania Path BioResource Molecular Profiling Core for Affymetrix GeneChip Mouse Gene 2.0 ST analysis.
Fetal Brain RNA-Seq
Illumina single-end messenger RNA (mRNA)-Seq libraries were prepared from 1 μg of E12.5 whole-brain RNA using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s protocol. Adaptors containing unique nucleotide indexes were ligated to purified double-stranded cDNA and amplified for 10 cycles. Libraries were diluted to 1.8 pM and sequenced on the Illumina NextSeq 500 system. On average, 41 million reads were obtained per library. Raw data were aligned to the Mus musculus (GRCm38) reference genome with Rsubread (41). Counts were normalized to library size prior to differential expression analysis.
Statistics
All analyses were conducted by an investigator blinded to genotype. Phenotypic assessments of adult physiology and behavior were analyzed by two-way analysis of variance or multivariate analysis of variance for sex and genotype, with repeated measures where appropriate using JMP11 Pro software (SAS, Cary, NC). Where basal sex differences were detected, genotype effects were subsequently analyzed within sex. Main effects and interactions were further analyzed by Fisher’s protected least significant difference test. Values greater than 2 standard deviations from the group mean were removed from analysis. Significance was set at p < .05. Robust multi-array–normalized microarray data were analyzed by gene set enrichment analysis (GSEA; Broad Institute, Cambridge, MA) for changes in c2_CP, c5_BP, and c3 gene sets from the Molecular Signature Database (MsigDB, v4.0, Broad Institute) using 1000 gene set permutations and stringent significance criteria (normalized enrichment score > 1.8, false discovery rate [FDR] < 0.05) (42,43). Normalized RNA-Seq count data were assessed for differential expression using the DESeq package in R Bioconductor (44). Genes with FDR < 0.05 and p < .05 were considered differentially expressed. Functional annotation clustering of differentially expressed genes was performed using Database for Annotation, Visualization, and Integrated Discovery tools, where clusters with enrichment scores > 1.3 and FDR < 0.05 were considered significant (45). Heat maps were generated in R using gplots.
RESULTS
Placental Trophoblast-Specific InsR Deletion Preserves Viability, Growth, and Metabolic Function
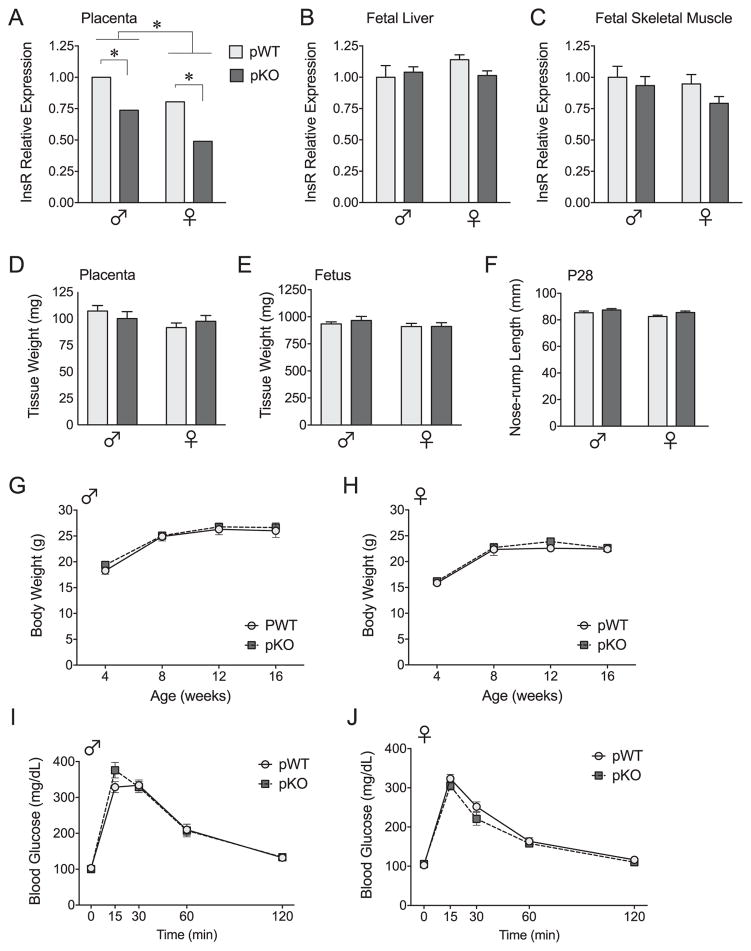
Effective InsR targeting was confirmed by quantitative reverse transcription polymerase chain reaction (Figure 1A) and Western blot (Supplemental Figure S1A) on E12.5, after full differentiation of the placenta, in pKO conceptuses and their littermate controls (36). InsR mRNA was significantly reduced in male and female pKO placentas at a magnitude expected based on the presence of maternal decidua and endothelial cells. InsR expression remained unaltered in the fetal and adult brain (Supplemental Figure S1B, C) and in insulin-responsive liver and skeletal muscle samples at E17.5 (Figure 1B, C). Placental InsR deletion did not affect viability with the observed genotypes, χ2(5, N = 151) = 3.26, p = .66, and sex ratio, χ2(1, N = 151) = 0.06, p = .81, not differing from expected Mendelian inheritance ratios.
Figure 1.
Placental-specific insulin receptor (InsR) targeting preserves growth and metabolic function. (A–C) Effective InsR targeting in fetally derived placental trophoblasts was confirmed by quantitative reverse transcription polymerase chain reaction in placental trophoblast-specific InsR deficiency (pKO) conceptuses and their littermate controls (pWT). (A) As expected, in the fully differentiated embryonic day (E) 12.5 placenta (n = 4/group), InsR messenger RNA (mRNA) was significantly reduced in pKO male and female mice, genotype: F1,11 = 47.21, p < .0001. Male mice had higher InsR expression in the placenta at this gestational stage, sex: F1,11 = 27.64, p = .0003; however, the effect of sex did not interact with genotype, interaction: F1,11 = 0.38, p = .55. (B, C) Confirming tissue-specific InsR targeting, InsR mRNA remained unaltered in fetal insulin-responsive tissues, including liver, genotype: F1,16 = 0.54, p = .47, interaction: F1,16 = 2.056, p = .17, and skeletal muscle, genotype: F1,14 = 2.11, p = .17, interaction: F1,14 = 0.34, p = .57, at E17.5 (n = 5/group). (D, E) No differences in preparturition placenta weights, genotype: F1,34 = 0.012, p = .91, interaction: F1,34 = 1.42, p = .24, and fetus weights, genotype: F1,34 = 0.30, p = .59, interaction: F1,4 = 0.27, p = .61, were detected at E17.5 (n = 7–12 litters/group, weights averaged within litter). (F) In pKO mice (n = 8–10/group), there was a main effect of genotype on body length at weaning, F1,30 = 4.83, p = .036; however, no within-sex differences were detected by Fisher’s protected least significant difference test. (G, H) Analysis of body weight in these mice at 4, 8, 12, and 16 weeks of age indicated no impact of placental InsR deletion on growth across the lifespan of male mice, genotype: F1,16 = 0.27, p = .61, genotype × age: F3,48 = 0.32, p = .81, or female mice, genotype: F1,10 = 2.81, p = .12, genotype × age: F3,30 = 0.80, p = .50. (I, J) As further confirmation that metabolic processes were intact, pKO male and female mice showed no differences in their rate of glucose clearance as fasted adults, genotype: F1,42 = 0.057, p = .81, genotype × sex: F1,42 = 1.60, p = .21, genotype × sex × time: F4,39 = 1.69, p = .17. Values are mean ± SEM. *p < .05.
To confirm that neurodevelopmental outcomes were not attributable to gross metabolic effects of placental-specific InsR deletion, we assessed preparturition tissue weights, postnatal growth, and glucose tolerance. Consistent with the limited effects of global InsR knockout on prenatal growth (46), there were no effects of genotype on E17.5 placenta or fetus weights (Figure 1D, E). At weaning, we detected a main effect of genotype on nose–rump length, where post hoc tests revealed nonsignificant increases in body length of pKO male and female mice of 2.4% and 3.5%, respectively (Figure 1F). However, there were no differences in body weight across the lifespan or rate of glucose clearance in pKO mice (Figure 1G–J).
Placental InsR Deficiency Recapitulates Sex-Biased Endophenotypes of Neurodevelopmental Dsorders
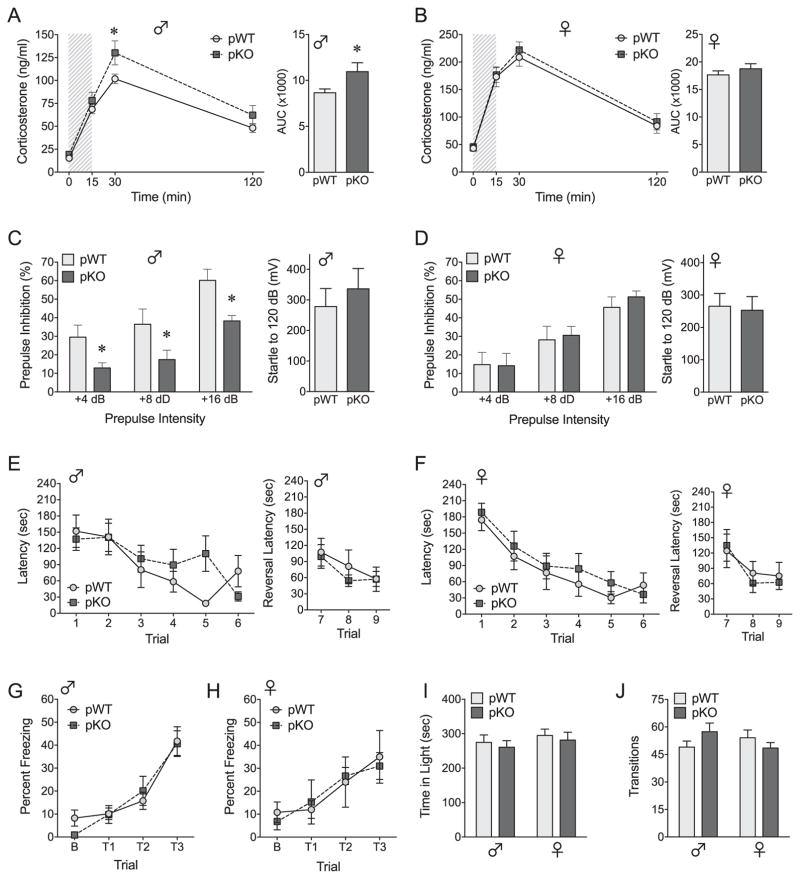
Adult pKO mice and their littermate controls were compared in translatable outcome measures of stress sensitivity, sensorimotor gating, and cognition, functional domains affected in sex-biased disorders, including autism, ADHD, and schizophrenia (47–49). In the HPA stress axis assessment, acute restraint increased plasma corticosterone in all groups (time: F3,41 = 264.07, p < .0001) and to a greater extent in females as expected (sex: F1,43 = 114.47, p < .0001; sex × time: F3,41 = 24.78, p < .0001). Subsequent within-sex analyses revealed significantly increased corticosterone responses to restraint in pKO male mice (Figure 2A) but no genotype effects in female mice (Figure 2B). In our assessment of sensorimotor gating, placental InsR deficiency induced sex-specific impairment of PPI. Despite normal baseline startle reflexivity, the ability of a weak prepulse to reduce or “gate” the response to a subsequent startling stimulus was profoundly disrupted at all prepulse intensities in pKO male mice (Figure 2C) yet remained intact in pKO female mice (Figure 2D).
Figure 2.
Placental insulin receptor (InsR) deficiency recapitulates male-specific endophenotypes of neurodevelopmental disorders. (A, B) Time course and area under the curve (AUC) analyses of corticosterone responses to a 15-minute restraint (shaded region) depict male-specific effects of placental InsR deletion on hypothalamic-pituitary-adrenal (HPA) axis responsiveness. (A) In adult male mice, total corticosterone over 120 minutes was significantly elevated in placental trophoblast-specific InsR deficiency (pKO) mice relative to controls (pWT), AUC: t21 = 2.22, p = .037. The effect of pKO manifested as a significant increase in peak corticosterone secretion at 30 minutes, t21 = 2.75, p = .0073, although multivariate analysis of variance detected only a trending effect of pKO across time, genotype: F1,21 = 4.09, p = .055, genotype × time: F3,19 = 2.22, p = .12. (B) In female mice, pKO had no impact on corticosterone secretion, genotype: F1,22 = 0.43, p = .52, genotype × time: F3,20 = 0.18, p = .91, AUC: t22 = 0.96, p = .35. (C, D) Placental InsR deficiency also elicited a male-specific impairment of prepulse inhibition (PPI), genotype × sex: F1,36 = 4.15, p = .049, without affecting baseline acoustic startle responses to 120 dB, genotype: F1,36 = 0.19, p = .66, genotype × sex: F1,36 = 0.45, p = .51. (C) In adult male mice, PPI was reduced in pKO mice relative to pWT mice overall (p = .018) and at all prepulse intensities (+4 dB: p = .0005; +8 dB: p = .0001; +16 dB: p < .0001). (D) In contrast, there was no effect of genotype on PPI in female mice overall (p = .73) or at any prepulse intensity (+4 dB: p = .91; +8 dB: p = .60; +16 dB: p = .23). (E–J) Behavioral effects of placental InsR deficiency in adult male mice were selective, with pKO having no impact on cognitive or locomotor measures. (E, F) Spatial memory acquisition (trials 1–6) and reversal learning (trials 1–9) in the Barnes maze were unaltered in pKO adult male mice (E) and female mice (F). Latency to find the target escape box improved across the 6 acquisition trials in all groups, F5,30 = 19.094, p < .0001, and there were no effects of sex, F1,34 = 0.004, p = .98, or genotype, F1,34 = 1.14, p = .29, trial × sex × genotype: F5,30 = 0.99, p = .44. Similarly, reversal latency (target rotated 180°) improved across trials, F2,33 = 6.14, p = .0054, and no effects of sex, F1,34 = 0.66, p = .42, or genotype, F1,34 = 0.32, p = .58, genotype × sex F2,33 = 0.14, p = .87, were detected. (G, H) Acquisition of auditory fear conditioning, depicted as percentage freezing to the tone (conditioned stimulus), increased similarly in all groups in response to three tone–shock pairings, trial × sex × genotype: F2,23 = 0.37, p = .69, and no main effect of genotype was observed, F1,24 = 1.57, p = .22. (I, J) In the light–dark box exploration test, there were no genotype effects on the amount of time spent in the light zone of the apparatus, genotype: F1,48 = 0.43, p = .52, genotype × sex: F1,48 = 0.00014, p = .99, or the number of light–dark zone transitions, genotype: F1,48 = 0.12, p = .73. Whereas we observed a trending interaction between the effects of sex and genotype on transitions, F1, 48 = 3.12, p = .084, no pairwise differences were detected (male pWT vs. pKO: p = .16; female pWT vs. pKO: p = .30). Values are mean ± SEM. HPA, PPI, and light/dark cohort: n = 9–14/group. Barnes maze and fear conditioning cohort: n = 8–10/group. *p < .05.
The phenotypic effects of placental InsR deficiency in male mice were highly specific, with pKO male and female mice exhibiting intact spatial memory acquisition and reversal learning in the Barnes maze (Figure 2E, F), normal auditory fear conditioning (Figure 2G, H), and similar avoidance of stress-provoking stimuli in the light–dark exploration test (Figure 2I). Furthermore, no differences in locomotor activity were detected (Figure 2J).
Long-Term PFC Reprogramming in Male Mice With Placental Trophoblast-Specific InsR Deficiency
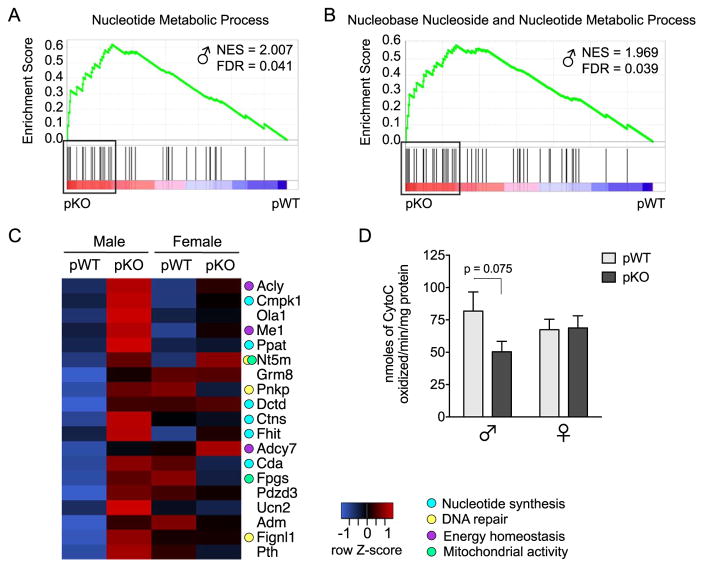
The PFC modulates both HPA axis activity and PPI in rodents, and its sex-specific susceptibility to early life challenge has been well established (50–55). To identify PFC changes contributing to phenotypic effects in these mice, PFC gene expression profiles were determined by genome-wide microarray. GSEA of the PFC transcriptome identified two gene sets differentially expressed in pKO versus pWT male mice and no altered gene sets in female mice (Table 1). In pKO male mice, gene sets for nucleotide metabolic processes were increased relative to pWT (Figure 3A, B). Analysis of the leading-edge subset, defined as the gene set members accounting for the enrichment, showed upregulation of genes related to nucleotide synthesis, DNA repair, metabotropic glutamate receptor signaling, and mitochondrial activity in pKO male mice (Figure 3C). To directly assess mitochondrial function, we measured cytochrome C oxidase (CCO) activity in the PFC of test-naïve adults (Figure 3D). Whereas we detected lower CCO activity in pKO males, the effects of genotype and sex did not reach statistical significance.
Table 1.
Summary of PFC Gene Sets Altered by InsR Deletion
| Type | Total | Male PFC
|
Female PFC
|
||
|---|---|---|---|---|---|
| Suppressed | Induced | Suppressed | Induced | ||
| c2.cp | 1019 | 0 | 0 | 0 | 0 |
|
| |||||
| c5.bp | 576 | 0 | 2 | 0 | 0 |
|
| |||||
| c3.all | 775 | 0 | 0 | 0 | 0 |
Number of gene sets significantly induced or suppressed in placental trophoblast-specific InsR deficiency (pKO) PFC relative to controls (pWT) (normalized enrichment score > 1.8 and false discovery rate < 0.05).
InsR, insulin receptor; PFC, prefrontal cortex.
Figure 3.
Long-term prefrontal cortex (PFC) programming in male mice with placental insulin receptor (InsR) deficiency points to a disruption in cellular metabolic processes. (A, B) Gene set enrichment analysis (GSEA) of the adult PFC transcriptome in placental trophoblast-specific InsR deficiency (pKO) vs. control (pWT) male mice (n = 6/group) identified two significantly enriched gene sets in the pKO group, indicating programming of PFC nucleotide/nucleoside metabolic processes. Plots depict gene set enrichment score, normalized enrichment score (NES), distribution of set members across the GSEA-ranked gene list, and genes comprising the leading-edge subset (boxes). (C) Leading-edge analysis revealed an upregulation of genes involved in nucleotide synthesis, DNA repair, metabotropic glutamate receptor signaling, and mitochondrial activity in the PFC of adult pKO male mice. The heat map depicts expression of the leading-edge subset in the adult PFC, where blue indicates lower and red indicates higher average relative gene expression across groups. (D) Cytochrome C oxidase (CCO) activity in the PFC of experiment-naïve adults. As would be predicted from the GSEA results, a trending decrease in CCO activity was detected in pKO male mice (a priori t test, pWT vs. pKO male mice: t13 = 1.94, p = .075); however, these results did not reach statistical significance, interaction: F1,2 = 2.61, p = .12, genotype: F1,27 = 2.20, p = .15, sex: F1,27 = 0.039, p = .85. n = 7–8/group. FDR, false discovery rate.
InsR Deficiency Induces a Transcriptional Response Predominantly in Male Placentas
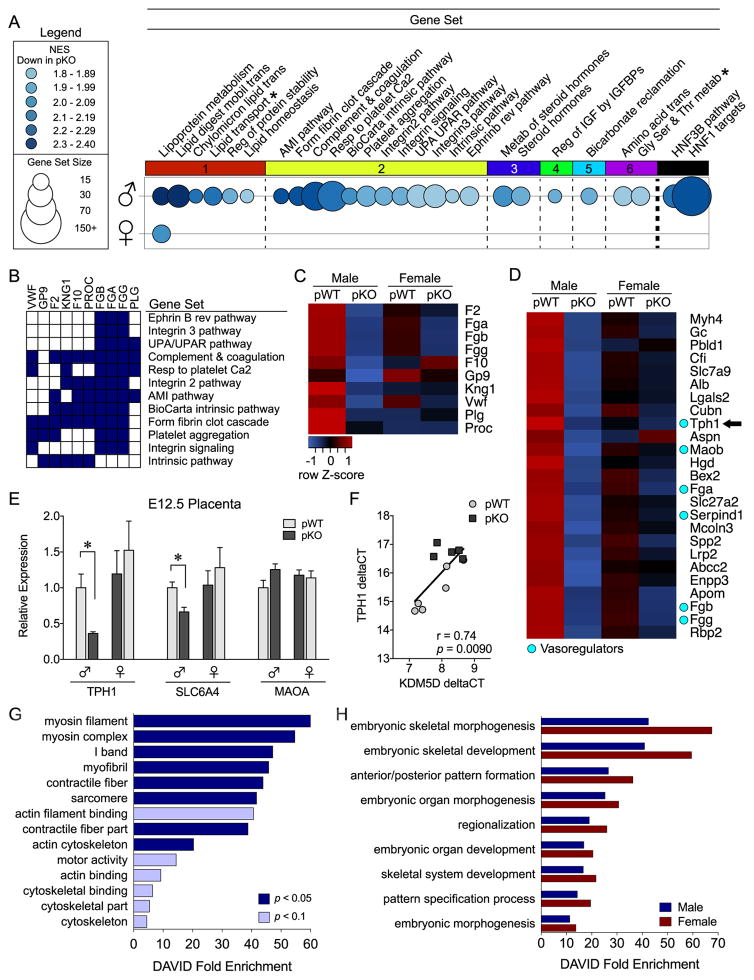
To identify mechanisms driving sex-specific reprogramming, we evaluated gene expression profiles in late-gestation (E17.5) placentas by genome-wide microarray and GSEA. InsR deficiency suppressed 26 gene sets in male placentas and 1 gene set in female placentas (Figure 4A). No gene sets were significantly upregulated in pKO placentas (Table 2). The maximally repressed gene set in male mice, Reactome_Lipoprotein_Metabolism, was the only gene set affected in pKO female mice (Supplemental Tables S2 and S3). Furthermore, placental cholesterol transport and triglyceride metabolic genes comprised the leading-edge subset and were suppressed by InsR deletion in both sexes (Supplemental Figure S2A–C). Consistent with these transcriptional effects, levels of cholesteryl esters and triglycerides in the E17.5 brain were similarly altered in pKO male and female mice (Supplemental Figure S2D–G).
Figure 4.
Insulin receptor (InsR) deficiency elicits sex-specific transcriptional responses in the placenta and fetal brain. (A–C) Gene set enrichment analysis of embryonic day (E) 17.5 placenta transcriptome identified key pathways dysregulated by InsR deletion (n = 6/group). (A) Bubble plot of gene sets (circles) significantly downregulated in male (top) and female (bottom) placental trophoblast-specific InsR deficiency (pKO) placentas relative to controls (pWT), where color intensity indicates the normalized enrichment score (NES) and diameter indicates gene set size. Differentially expressed gene sets (NES > 1.8, false discovery rate [FDR] < 0.05) were clustered by gene member overlap and ordered by NES. Whereas InsR deletion suppressed lipoprotein metabolism genes in both sexes, 25 additional gene sets were downregulated only in male pKO placentas. Clustering illustrated marked suppression of genes mediating (1) lipid homeostasis, (2) vascular function, (3) steroid hormone metabolism, (4) insulin-like growth factor (IGF) activity, (5) mitochondrial function, and (6) amino acid transport in pKO male mice. Gene set enrichment analysis (GSEA) also identified hepatocyte nuclear factor 1 (HNF1) as a candidate regulator in male mice, with the majority of gene sets containing HNF1 targets (* = not HNF1 target). Differentially expressed gene sets are detailed in Supplemental Table S2. (B) Plot showing the overlap of the leading-edge genes from each differentially expressed gene set in the vascular cluster. Expression of a common subset of coagulation cascade genes was suppressed in pKO male placentas. (C) A heat map of genes comprising the coagulation subset, where blue indicates lower and red indicates higher average relative expression across groups. InsR deletion decreased expression of procoagulant genes in male mice. (D) A heat map of the top 20 GSEA-ranked genes correlated with InsR deletion in male E17.5 placentas. Cyan circles indicate vasoregulators, including the rate-limiting serotonin synthesizing enzyme tryptophan hydroxylase 1 (TPH1, arrow). (E) Consistent with E17.5, TPH1 messenger RNA was suppressed by InsR knockout by E12.5 in male placentas (a priori t test, pWT vs. pKO male mice: t9 = 3.0, p = .015) but not in female mice (t10 = 0.63, p = .54), although the interaction between genotype and sex did not reach statistical significance, F1,19 = 2.75, p = .11. The serotonin reuptake transporter (SLC6A4) was similarly decreased in male tissue (a priori t test, pWT vs. pKO male mice: t9 = 3.25, p = .0087) but not in female mice (t10 = 0.71, p = .49), whereas no genotype by sex interaction was detected, F1,20 = 2.61, p = .12. Conversely, placental expression of monoamine oxidase A (MAOA) in male mice was not significantly affected by InsR deficiency (a priori t test, pWT vs. pKO male mice: t9 = 1.87, p = .095). (F) Expression of the Y-linked histone demethylase KDM5D was positively correlated with TPH1 in male E12.5 placentas (R2 9 = .55, p = .009). (G–H) Because placenta-derived serotonin is critical for fetal neurodevelopment, we assessed the E12.5 brain transcriptome for sex-specific effects of pKO. Functional annotation clustering of differentially expressed genes suggested disruption of motility and cytoskeletal dynamics only in pKO male mice (G), whereas the remaining effects of pKO were detected in both male and female mice (H). AMI, acute myocardial infarction; DAVID, Database for Annotation, Visualization, and Integrated Discovery; HNF3B, hepatocyte nuclear factor 3 beta; UPA, urokinase-type plasminogen activator; UPAR, UPA receptor.
Table 2.
Summary of placental gene sets altered by InsR deletion
| Type | Total | Male Placenta
|
Female Placenta
|
||
|---|---|---|---|---|---|
| Suppressed | Induced | Suppressed | Induced | ||
| c2.cp | 1019 | 22 | 0 | 1 | 0 |
|
| |||||
| c5.bp | 576 | 3 | 0 | 0 | 0 |
|
| |||||
| c3.all | 776 | 1 | 0 | 0 | 0 |
Number of gene sets significantly induced or suppressed in placental trophoblast-specific InsR deficiency (pKO) embryo day 17.5 placentas relative to controls (pWT) (false discovery rate < 0.05 and normalized enrichment score > 1.8).
InsR, insulin receptor.
In male placentas, InsR deficiency suppressed 25 additional gene sets involving lipid homeostasis, vascular function, steroid hormone metabolism, insulin-like growth factor activity, mitochondrial function, and amino acid transport/metabolism (Figure 4A and Supplemental Table S2). The transcription factor, hepatocyte nuclear factor 1, was identified as a candidate regulator together with HNF3B, as 92% of affected gene sets contained suppressed hepatocyte nuclear factor 1 targets. Based on the pleotropic influence of the placental vasculature, we further examined the male-specific altered vascular profile. Leading-edge analysis revealed that a subset of coagulation genes accounted for the cluster’s suppression (Figure 4B, C). InsR deficiency promoted an anticoagulant profile in male placentas, including decreased fibrinogen, thrombin, von Willebrand factor, kinogen 1, and factor X expression (Figure 4C). Furthermore, vasoregulators were among the top 20 GSEA-ranked genes, including decreased fibrinogen, heparin cofactor 1, monoamine oxidase B, and the rate-limiting serotonin synthesizing enzyme tryptophan hydroxylase 1 (TPH1) (Figure 4D).
The Y-linked H3K4me3 Demethylase, KDM5D, Predicts Serotonergic Gene Expression in Male InsR-Deficient Placentas
We hypothesized that this anticoagulant profile may be related to intrauterine levels of serotonin (5–hydroxytryptamine), a potent vasoconstrictor and essential regulator of fetal neurodevelopment, particularly during midgestation (56–58). As such, we next assessed for sex-specific effects of InsR deficiency on serotonin-related gene expression in E12.5 placenta hemisections (Figure 4E). Consistent with E17.5, InsR deficiency decreased expression of TPH1 at E12.5 in male mice only. Expression of the serotonin reuptake transporter (SLC6A4/SERT) was also reduced in pKO male mice. No differences in monoamine oxidase A expression were observed. To elucidate molecular mechanisms mediating male-specific placental serotonergic effects, we examined the relationship between the expression of TPH1 and sex chromosome–linked placental epigenetic regulators, including the H3K4me3 demethylases, KDM5C (X linked) and KDM5D (Y linked), and the H3K27me3 demethylases, UTX and UTY. We detected a significant positive correlation between placental TPH1 and KDM5D expression in male mice, thereby identifying this Y-linked histone demethylase as a candidate epigenetic mediator (Figure 4F). No correlations were detected for KDM5C, UTX, or UTY (data not shown).
We next assessed levels of biogenic amines in the remaining placenta hemisections by liquid chromatography/mass spectrometry. Although we detected an interaction between genotype and sex on placental serotonin levels (interaction F1,20 = 4.54, p = .046, post hoc tests did not reach significance (Supplemental Figure S3A). No differences were detected in levels of 5-hydroxyindoleacetic acid, the major metabolite of serotonin (Supplemental Figure S3B). Whereas we observed sex differences in dopamine and norepinephrine levels, no genotype effects were detected (Supplemental Figure S3C, D).
Sex-Specific Changes in the Fetal Brain Transcriptome in Response to Placental InsR Deficiency
Next, we used RNA-Seq to determine whether placental serotonergic changes at E12.5 were associated with dysregulated neurodevelopment in pKO male mice, as predicted based on the involvement of serotonin in fetal brain cell proliferation, migration, and wiring (58,59). In total, 27 and 32 genes were significantly differentially expressed in the E12.5 brain of male and female pKO mice, respectively (Supplemental Table S4). Notably, 13 genes were affected by InsR deficiency in both sexes, including a subset of homeobox transcription factors involved in brainstem dorsal–ventral patterning. Gene ontology analysis of the genes significantly dysregulated by InsR deficiency using Database for Annotation, Visualization, and Integrated Discovery tools revealed an overrepresentation of genes mediating cytoskeletal dynamics and motility only in pKO males (Figure 4G). The remaining gene ontology terms were similarly affected in male and female mice (Figure 4H) and reflected changes in dorsal–ventral patterning and embryonic morphogenesis.
DISCUSSION
Diabetes, obesity, and overweight are prevalent pregnancy complications that predispose offspring to male-biased neurodevelopmental disorders, including ADHD, autism, and schizophrenia (1–8). Whereas the mechanisms leading to sex differences in disease risk remain unidentified, defective placental insulin signaling is common to these pregnancy complications and has been implicated in adverse fetal outcomes, especially in male individuals (19–28). We hypothesized that impaired insulin signaling in the placenta, the critical and sex-specific maternal–fetal intermediary, is a mechanistic link between maternal metabolic dysfunction and male-biased neurodevelopmental reprogramming. In the current study, to demonstrate the precise role of placental InsRs, end points related to neurodevelopmental disorders were evaluated in mice with placental trophoblast-specific InsR deletion and subsequent programmatic mediators were identified.
Excitingly, and consistent with our hypothesis, pKO produced endophenotypes of neurodevelopmental disorders in male mice but not in female mice. Importantly, our initial analyses confirmed that behavioral outcomes were independent of potential confounds related to offspring growth or metabolic dysfunction given that we found no changes in placental weight, fetal weight, longitudinal growth, or glucose tolerance with pKO. Impaired prenatal growth was not expected in the current study because insulin-like growth factor receptors have been shown to maintain placental and fetal growth in total InsR knockout mice (46). The sex specificity of these findings is remarkable and highlights the novel importance of placental function in neurodevelopmental programming. In examination of programmed stress reactivity in these mice, adult pKO male mice exhibited an increased HPA stress axis response, a common feature underlying neurodevelopmental and affective disorders (60–63). Adult pKO male mice also exhibited impaired sensorimotor gating (PPI), recapitulating abnormalities characteristic of neurodevelopmental disorders, including autism, ADHD, and schizophrenia. Such outcomes have significant translational value because both HPA axis stress reactivity and PPI are reproducible across species, including in humans, where the regulatory circuits are broadly conserved (51,55,64).
These neural stress circuits converge in the PFC, a critical regulator of both neuroendocrine stress responses and PPI and a brain region with known sensitivity to prenatal adversity that is disrupted in neurodevelopmental disorders (50–54,65). Therefore, we performed GSEA of the adult PFC transcriptome to identify molecular pathways dysregulated by pKO and associated with the male-specific phenotype. Placental trophoblast-specific InsR deletion resulted in programming of PFC gene expression in male mice but not in female mice. Whereas no differentially expressed gene sets were detected in female mice, gene sets for nucleotide metabolic processes, including nucleotide synthesis, nuclear and mitochondrial DNA repair, and cellular energy homeostasis, were increased in the PFC of pKO male mice. Surprisingly, we also observed increased expression of the immunomodulatory neuropeptides, adrenomedullin and urocortin 2, likely by local immune cells within the PFC (66–68). Together, these changes suggest an ongoing response to injury, oxidative stress, and/or mitochondrial dysfunction in the PFC of pKO male mice (69). Because our assessment of mitochondrial CCO activity in a separate test-naïve cohort did not reach statistical significance, the transcriptomic profile in behavior-tested pKO male mice may reflect an exacerbated state and a programmed susceptibility of the PFC to environmental stressors. Such outcomes are consistent with the observed mitochondrial insufficiency in fetal cells of diabetic pregnancies, which may then be worsened by stress, especially in the PFC, as has been implicated in the pathogenesis of schizophrenia and autism (70–76).
Placental trophoblasts are fetally derived, express the fetal genetic sex, and exhibit basal sex differences; therefore, we hypothesized that InsR deficiency elicited distinct patterns in male and female placentas and, in turn, orchestrated divergent fetal developmental trajectories (30,35). To identify sex-specific placental responses, we leveraged GSEA to compare expression of key molecular pathways in the late-gestation placenta. Strikingly, and consistent with the observed phenotypic outcomes, the majority of placental differences occurred again only in male mice. Only male pKO placentas exhibited an anticoagulant profile and repression of genes mediating amino acid metabolism and transport, steroid hormone synthesis, insulin-like growth factor activity, and mitochondrial function. In agreement with our results, sex differences in placental responses to intrauterine challenges in humans, including maternal diabetes and overnutrition, have been well established (28,29,35,77). InsR deficiency disrupted placental lipoprotein metabolism and cholesterol transfer similarly in male and female mice, recapitulating findings reported in human and rat diabetic placentas, yet likely having limited neural impact due to fetal de novo lipid synthesis at this gestational stage (19,78–82).
In examining the molecular pathways in the male placenta uniquely positioned to promote neurodevelopmental changes, regulation of serotonin production stood out as particularly important based on its known involvement in neurodevelopmental programming (58). In male mice, InsR deficiency suppressed genes involved in placental synthesis and clearance of serotonin, a potent vasoconstrictor implicated in the etiology of gestational diabetes (83). Similar serotonergic alterations in diabetic placentas have been directly attributed to impaired insulin signaling and undoubtedly affect neurogenesis, migration, and synaptogenesis as well as uteroplacental blood flow (20,56–58,83). Given the critical role of placenta-derived serotonin in forebrain development prior to E14.5, we assessed the effect of InsR deficiency on serotonergic gene expression and monoamine levels in the E12.5 placenta (84). Similar to our findings in late gestation, InsR deficiency suppressed TPH1 and SERT expression in male mice but not in female mice on E12.5; however, our assessment of serotonin levels did not reach statistical significance and was limited by the presence of maternally derived decidual and blood serotonin. To identify molecular mechanisms driving the male specificity of these findings, we evaluated expression of X- and Y-linked histone demethylases, epigenetic machinery capable of programming longterm changes in gene expression in a sex-dependent manner (85,86). In this examination, we identified the Y-linked histone demethylase, KDM5D, as a potential epigenetic mediator based on its positive correlation with TPH1 expression. Given that KDM5D demethylates lysine 4 of histone H3, thereby removing an activating histone mark, KDM5D activity may underlie repressed placental TPH1 in pKO male mice.
We predicted that these sex-specific placental responses to InsR would result in divergent neurodevelopmental trajectories in the developing fetal brain. Specifically, based on the known involvement of placental-derived serotonin in fetal brain development, we predicted changes related to cell migration, proliferation, and/or wiring in the pKO male brain (58). Therefore, we determined gene expression profiles in the E12.5 brain corresponding to the same placental time point examined above. Whereas a subset of genes involved in hindbrain dorsal–ventral patterning was altered by pKO in both sexes, only the pKO male mice exhibited reduced expression of genes modulating cytoskeletal dynamics and cell motility. These processes are required for normal neuronal proliferation, migration, and axon targeting during this gestational stage (87–89). Thus, these findings may reflect delayed or disrupted cortical development specifically in pKO male mice. In line with this, neonates born to diabetic humans and rodents exhibit markers of delayed brain and dendritic maturation (3,90). These data suggest a potential mechanism by which placental trophoblast-specific InsR deletion leads to long-term programmatic effects specifically in male mice.
The current study aimed to delineate the specific role of InsR in placental trophoblasts, the cell population known to exhibit insulin resistance in human diabetic placental tissue (21). Effects on InsR outside of trophoblast cells were not detected at E12.5, at E17.5, or in the adult brain; however, the potential impact at additional time points has not been examined. In addition, placental and fetal brain samples were composed of heterogeneous cell types, and therefore potential cell-type or region-specific effects of InsR deficiency may be undetected in the embryonic analyses presented here.
CONCLUSIONS
These studies demonstrate that perturbation of InsRs specifically in placental trophoblasts, a known consequence of maternal metabolic conditions that increases neurodevelopmental disorder risk, is sufficient to disrupt stress sensitivity and prefrontal cortical development in male mice but not in female mice. These findings provide compelling evidence for the critical involvement of placental InsRs in sex-specific neurodevelopment and have important implications for pregnancies complicated by diabetes and obesity.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants Nos. MH091258, MH073030, MH099910, MH087597, MH104184, and MH108286.
We thank Gustavo Leone at The Ohio State University for generous donation of the CYP19-Cre mice, Dan Beiting for technical assistance with transcriptomics analyses, Satish Srinivasan and Narayan Avadhani for assistance with the CCO assay, Itzhak Nissim at the University of Pennsylvania Children’s Hospital of Philadelphia Metabolomics core for biogenic amine quantification, Joshua Rabinowitz and Wenyun Lu at Princeton University for lipid quantification, and the University of Pennsylvania Diabetes Research Center for the use of the metabolomics core (P30-DK19525).
Footnotes
DISCLOSURES
The authors all report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2016.12.025.
References
- 1.Torres-Espinola FJ, Berglund SK, García-Valdés LM, Segura MT, Jerez A, Campos D, et al. Maternal obesity, overweight and gestational diabetes affect the offspring neurodevelopment at 6 and 18 months of age—A follow-up from the PREOBE cohort. PLoS One. 2015;10:e0133010. doi: 10.1371/journal.pone.0133010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ornoy A, Ratzon N, Greenbaum C, Wolf A, Dulitzky M. School-age children born to diabetic mothers and to mothers with gestational diabetes exhibit a high rate of inattention and fine and gross motor impairment. J Pediatr Endocrinol Metab. 2001;14(Suppl 1):681–689. doi: 10.1515/jpem.2001.14.s1.681. [DOI] [PubMed] [Google Scholar]
- 3.Ornoy A. Growth and neurodevelopmental outcome of children born to mothers with pregestational and gestational diabetes. Pediatr Endocrinol Rev. 2005;3:104–113. [PubMed] [Google Scholar]
- 4.Van Lieshout RJ, Voruganti LP. Diabetes mellitus during pregnancy and increased risk of schizophrenia in offspring: A review of the evidence and putative mechanisms. J Psychiatry Neurosci. 2008;33:395–404. [PMC free article] [PubMed] [Google Scholar]
- 5.Jo H, Schieve LA, Sharma AJ, Hinkle SN, Li R, Lind JN. Maternal prepregnancy body mass index and child psychosocial development at 6 years of age. Pediatrics. 2015;135:e1198–e1209. doi: 10.1542/peds.2014-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sells CJ, Robinson NM, Brown Z, Knopp RH. Long-term developmental follow-up of infants of diabetic mothers. J Pediatr. 1994;125:S9–S17. doi: 10.1016/s0022-3476(94)70170-9. [DOI] [PubMed] [Google Scholar]
- 7.Khandaker GM, Dibben CRM, Jones PB. Does maternal body mass index during pregnancy influence risk of schizophrenia in the adult offspring? Obes Rev. 2012;13:518–527. doi: 10.1111/j.1467-789X.2011.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera HM, Christiansen KJ, Sullivan EL. The role of maternal obesity in the risk of neuropsychiatric disorders. Front Neurosci. 2015;9:194. doi: 10.3389/fnins.2015.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huda SS, Brodie LE, Sattar N. Obesity in pregnancy: Prevalence and metabolic consequences. Semin Fetal Neonatal Med. 2010;15:70–76. doi: 10.1016/j.siny.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Hunt KJ, Schuller KL. The increasing prevalence of diabetes in pregnancy. Obstet Gynecol Clin North Am. 2007;34:173–199. doi: 10.1016/j.ogc.2007.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSisto CL, Kim SY, Sharma AJ. Prevalence estimates of gestational diabetes mellitus in the United States: Pregnancy Risk Assessment Monitoring System (PRAMS), 2007–2010. Prev Chronic Dis. 2014;11:E104. doi: 10.5888/pcd11.130415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom B, Cohen RA, Freeman G. Summary health statistics for U.S. children: National Health Interview Survey, 2010. Vital Health Stat. 2011;10:1–80. [PubMed] [Google Scholar]
- 13.Autism and Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. [Accessed May 10, 2016];Prevalence of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States. 2010 Available at: http://www.cdc.gov/mmwr/preview/mmwrhtml/ss6302a1.htm. [PubMed]
- 14.Giraudo SQ, Della-Fera MA, Proctor L, Wickwire K, Ambati S, Baile CA. Maternal high fat feeding and gestational dietary restriction: Effects on offspring body weight, food intake and hypothalamic gene expression over three generations in mice. Pharmacol Biochem Behav. 2010;97:121–129. doi: 10.1016/j.pbb.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Kruse MS, Vega MC, Rey M, Coirini H. Sex differences in LXR expression in normal offspring and in rats born to diabetic dams. J Endocrinol. 2014;222:53–60. doi: 10.1530/JOE-14-0054. [DOI] [PubMed] [Google Scholar]
- 16.Dahlhoff M, Pfister S, Blutke A, Rozman J, Klingenspor M, Deutsch MJ, et al. Peri-conceptional obesogenic exposure induces sex-specific programming of disease susceptibilities in adult mouse offspring. Biochim Biophys Acta. 2014;1842:304–317. doi: 10.1016/j.bbadis.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Yokomizo H, Inoguchi T, Sonoda N, Sakaki Y, Maeda Y, Inoue T, et al. Maternal high-fat diet induces insulin resistance and deterioration of pancreatic β-cell function in adult offspring with sex differences in mice. Am J Physiol Endocrinol Metab. 2014;306:E1163–E1175. doi: 10.1152/ajpendo.00688.2013. [DOI] [PubMed] [Google Scholar]
- 18.Ornellas F, Mello VS, Mandarim-de-Lacerda CA, Aguila MB. Sexual dimorphism in fat distribution and metabolic profile in mice offspring from diet-induced obese mothers. Life Sci. 2013;93:454–463. doi: 10.1016/j.lfs.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Petropoulos S, Guillemin C, Ergaz Z, Dimov S, Suderman M, Weinstein-Fudim L, et al. Gestational diabetes alters offspring DNA methylation profiles in human and rat: Identification of key pathways involved in endocrine system disorders, insulin signaling, diabetes signaling, and ILK signaling. Endocrinology. 2015;156:2222–2238. doi: 10.1210/en.2014-1643. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Hadden C, Singh P, Mercado CP, Murphy P, Dajani NK, et al. GDM-associated insulin deficiency hinders the dissociation of SERT from ERp44 and down-regulates placental 5-HT uptake. Proc Natl Acad Sci U S A. 2014;111:E5697–E5705. doi: 10.1073/pnas.1416675112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Cooper A, Odibo IN, Ahmed A, Murphy P, Koonce R, et al. Discrepancy in insulin regulation between GDM-platelets and -placenta. J Biol Chem. 2016;291:9657–9665. doi: 10.1074/jbc.M116.713693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Colomiere M, Permezel M, Riley C, Desoye G, Lappas M. Defective insulin signaling in placenta from pregnancies complicated by gestational diabetes mellitus. Eur J Endocrinol. 2009;160:567–578. doi: 10.1530/EJE-09-0031. [DOI] [PubMed] [Google Scholar]
- 23.Desoye G, Hofmann HH, Weiss PA. Insulin binding to trophoblast plasma membranes and placental glycogen content in well-controlled gestational diabetic women treated with diet or insulin, in well-controlled overt diabetic patients and in healthy control subjects. Diabetologia. 1992;35:45–55. doi: 10.1007/BF00400851. [DOI] [PubMed] [Google Scholar]
- 24.Harrison LC, Billington T, Clark S, Nichols R, East I, Martin FI. Decreased binding of insulin by receptors on placental membranes from diabetic mothers. J Clin Endocrinol Metab. 1977;44:206–209. doi: 10.1210/jcem-44-1-206. [DOI] [PubMed] [Google Scholar]
- 25.Street ME, Viani I, Ziveri MA, Volta C, Smerieri A, Bernasconi S. Impairment of insulin receptor signal transduction in placentas of intra-uterine growth-restricted newborns and its relationship with fetal growth. Eur J Endocrinol. 2011;164:45–52. doi: 10.1530/EJE-10-0752. [DOI] [PubMed] [Google Scholar]
- 26.Rademacher TW, Gumaa K, Scioscia M. Preeclampsia, insulin signalling and immunological dysfunction: A fetal, maternal or placental disorder? J Reprod Immunol. 2007;76:78–84. doi: 10.1016/j.jri.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Scioscia M, Gumaa K, Kunjara S, Paine MA, Selvaggi LE, Rodeck CH, Rademacher TW. Insulin resistance in human preeclamptic placenta is mediated by serine phosphorylation of insulin receptor substrate-1 and −2. J Clin Endocrinol Metab. 2006;91:709–717. doi: 10.1210/jc.2005-1965. [DOI] [PubMed] [Google Scholar]
- 28.Bronson SL, Bale TL. The placenta as a mediator of stress effects on neurodevelopmental reprogramming. Neuropsychopharmacology. 2016;41:207–218. doi: 10.1038/npp.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myatt L. Placental adaptive responses and fetal programming. J Physiol. 2006;572:25–30. doi: 10.1113/jphysiol.2006.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson ED, Cross JC. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 31.Jansson T, Powell TL. Role of the placenta in fetal programming: Underlying mechanisms and potential interventional approaches. Clin Sci (Lond) 2007;113:1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- 32.O’Tierney-Ginn P, Presley L, Myers S, Catalano P. Placental growth response to maternal insulin in early pregnancy. J Clin Endocrinol Metab. 2015;100:159–165. doi: 10.1210/jc.2014-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desoye G, Hartmann M, Blaschitz A, Dohr G, Hahn T, Kohnen G, Kaufmann P. Insulin receptors in syncytiotrophoblast and fetal endothelium of human placenta: Immunohistochemical evidence for developmental changes in distribution pattern. Histochemistry. 1994;101:277–285. doi: 10.1007/BF00315915. [DOI] [PubMed] [Google Scholar]
- 34.Hiden U, Maier A, Bilban M, Ghaffari-Tabrizi N, Wadsack C, Lang I, et al. Insulin control of placental gene expression shifts from mother to foetus over the course of pregnancy. Diabetologia. 2006;49:123–131. doi: 10.1007/s00125-005-0054-x. [DOI] [PubMed] [Google Scholar]
- 35.Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C. Placental contribution to the origins of sexual dimorphism in health and diseases: Sex chromosomes and epigenetics. Biol Sex Differ. 2013;4:5. doi: 10.1186/2042-6410-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossant J, Cross JC. Placental development: Lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 37.Soares MJ, Chakraborty D, Karim Rumi MA, Konno T, Renaud SJ. Rat placentation: An experimental model for investigating the hemochorial maternal–fetal interface. Placenta. 2012;33:233–243. doi: 10.1016/j.placenta.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenzel PL, Leone G. Expression of Cre recombinase in early diploid trophoblast cells of the mouse placenta. Genesis. 2007;45:129–134. doi: 10.1002/dvg.20276. [DOI] [PubMed] [Google Scholar]
- 39.Bronson SL, Bale TL. Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology. 2014;155:2635–2646. doi: 10.1210/en.2014-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louvi A, Accili D, Efstratiadis A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev Biol. 1997;189:33–48. doi: 10.1006/dbio.1997.8666. [DOI] [PubMed] [Google Scholar]
- 47.Kas MJ, Glennon JC, Buitelaar J, Ey E, Biemans B, Crawley J, et al. Assessing behavioural and cognitive domains of autism spectrum disorders in rodents: Current status and future perspectives. Psychopharmacology (Berl) 2014;231:1125–1146. doi: 10.1007/s00213-013-3268-5. [DOI] [PubMed] [Google Scholar]
- 48.Powell CM, Miyakawa T. Schizophrenia-relevant behavioral testing in rodent models: A uniquely human disorder? Biol Psychiatry. 2006;59:1198–1207. doi: 10.1016/j.biopsych.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeler JF, Robbins TW. Translating cognition from animals to humans. Biochem Pharmacol. 2011;81:1356–1366. doi: 10.1016/j.bcp.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 50.Diorio D, Viau V, Meaney MJ. The role of the medial prefrontal cortex (cingulate gyrus) in the regulation of hypothalamic–pituitary–adrenal responses to stress. J Neurosci. 1993;13:3839–3847. doi: 10.1523/JNEUROSCI.13-09-03839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKlveen JM, Myers B, Herman JP. The medial prefrontal cortex: Coordinator of autonomic, neuroendocrine and behavioural responses to stress. J Neuroendocrinol. 2015;27:446–456. doi: 10.1111/jne.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radley JJ, Arias CM, Sawchenko PE. Regional differentiation of the medial prefrontal cortex in regulating adaptive responses to acute emotional stress. J Neurosci. 2006;26:12967–12976. doi: 10.1523/JNEUROSCI.4297-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selemon LD, Zecevic N. Schizophrenia: A tale of two critical periods for prefrontal cortical development. Transl Psychiatry. 2015;5:e623. doi: 10.1038/tp.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: Current knowledge and future challenges. Psychopharmacology (Berl) 2001;156:194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- 55.Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: Normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- 56.Velasquez JC, Goeden N, Bonnin A. Placental serotonin: Implications for the developmental effects of SSRIs and maternal depression. Front Cell Neurosci. 2013;7:47. doi: 10.3389/fncel.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riccio O, Potter G, Walzer C, Vallet P, Szabó G, Vutskits L, et al. Excess of serotonin affects embryonic interneuron migration through activation of the serotonin receptor 6. Mol Psychiatry. 2009;14:280–290. doi: 10.1038/mp.2008.89. [DOI] [PubMed] [Google Scholar]
- 58.Bonnin A, Levitt P. Fetal, maternal, and placental sources of serotonin and new implications for developmental programming of the brain. Neuroscience. 2011;197:1–7. doi: 10.1016/j.neuroscience.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonnin A, Torii M, Wang L, Rakic P, Levitt P. Serotonin modulates the response of embryonic thalamocortical axons to netrin-1. Nat Neurosci. 2007;10:588–597. doi: 10.1038/nn1896. [DOI] [PubMed] [Google Scholar]
- 60.Moghaddam B. Stress activation of glutamate neurotransmission in the prefrontal cortex: Implications for dopamine-associated psychiatric disorders. Biol Psychiatry. 2002;51:775–787. doi: 10.1016/s0006-3223(01)01362-2. [DOI] [PubMed] [Google Scholar]
- 61.Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 62.Walker E, Mittal V, Tessner K. Stress and the hypothalamic pituitary adrenal axis in the developmental course of schizophrenia. Annu Rev Clin Psychol. 2008;4:189–216. doi: 10.1146/annurev.clinpsy.4.022007.141248. [DOI] [PubMed] [Google Scholar]
- 63.Corbett BA, Schupp CW, Levine S, Mendoza S. Comparing cortisol, stress, and sensory sensitivity in children with autism. Autism Res. 2009;2:39–49. doi: 10.1002/aur.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell LE, Hughes M, Budd TW, Cooper G, Fulham WR, Karayanidis F, et al. Primary and secondary neural networks of auditory prepulse inhibition: A functional magnetic resonance imaging study of sensorimotor gating of the human acoustic startle response. Eur J Neurosci. 2007;26:2327–2333. doi: 10.1111/j.1460-9568.2007.05858.x. [DOI] [PubMed] [Google Scholar]
- 65.Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. J Neurosci. 2010;30:14595–14609. doi: 10.1523/JNEUROSCI.2257-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tixier E, Leconte C, Touzani O, Roussel S, Petit E, Bernaudin M. Adrenomedullin protects neurons against oxygen glucose deprivation stress in an autocrine and paracrine manner. J Neurochem. 2008;106:1388–1403. doi: 10.1111/j.1471-4159.2008.05494.x. [DOI] [PubMed] [Google Scholar]
- 67.Kempuraj D, Papadopoulou NG, Lytinas M, Huang M, Kandere-Grzybowska K, Madhappan B, et al. Corticotropin-releasing hormone and its structurally related urocortin are synthesized and secreted by human mast cells. Endocrinology. 2004;145:43–48. doi: 10.1210/en.2003-0805. [DOI] [PubMed] [Google Scholar]
- 68.Souza-Moreira L, Campos-Salinas J, Caro M, Gonzalez-Rey E. Neuropeptides as pleiotropic modulators of the immune response. Neuroendocrinology. 2011;94:89–100. doi: 10.1159/000328636. [DOI] [PubMed] [Google Scholar]
- 69.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 70.Batandier C, Poulet L, Hininger I, Couturier K, Fontaine E, Roussel A-M, Canini F. Acute stress delays brain mitochondrial permeability transition pore opening. J Neurochem. 2014;131:314–322. doi: 10.1111/jnc.12811. [DOI] [PubMed] [Google Scholar]
- 71.Kuchukashvili Z, Burjanadze G, Menabde K, Chachua M, Dachanidze N, Mikadze M, Koshoridze N. Long-lasting stress, quantitative changes in nitric oxide concentration and functional state of brain mitochondria. Acta Neurobiol Exp (Wars) 2012;72:40–50. doi: 10.55782/ane-2012-1879. [DOI] [PubMed] [Google Scholar]
- 72.Rezin GT, Amboni G, Zugno AI, Quevedo J, Streck EL. Mitochondrial dysfunction and psychiatric disorders. Neurochem Res. 2009;34:1021–1029. doi: 10.1007/s11064-008-9865-8. [DOI] [PubMed] [Google Scholar]
- 73.Dror N, Klein E, Karry R, Sheinkman A, Kirsh Z, Mazor M, et al. State-dependent alterations in mitochondrial complex I activity in platelets: A potential peripheral marker for schizophrenia. Mol Psychiatry. 2002;7:995–1001. doi: 10.1038/sj.mp.4001116. [DOI] [PubMed] [Google Scholar]
- 74.Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G, et al. Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia. Mol Neuropsychiatry. 2015;1:201–219. doi: 10.1159/000441252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim J, Piao Y, Pak YK, Chung D, Han YM, Hong JS, et al. Umbilical cord mesenchymal stromal cells affected by gestational diabetes mellitus display premature aging and mitochondrial dysfunction. Stem Cells Dev. 2015;24:575–586. doi: 10.1089/scd.2014.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lepagnol-Bestel A-M, Maussion G, Boda B, Cardona A, Iwayama Y, Delezoide A-L, et al. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Mol Psychiatry. 2008;13:385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- 77.Tarrade A, Panchenko P, Junien C, Gabory A. Placental contribution to nutritional programming of health and diseases: Epigenetics and sexual dimorphism. J Exp Biol. 2015;218:50–58. doi: 10.1242/jeb.110320. [DOI] [PubMed] [Google Scholar]
- 78.Dubé E, Ethier-Chiasson M, Lafond J. Modulation of cholesterol transport by insulin-treated gestational diabetes mellitus in human full-term placenta. Biol Reprod. 2013;88:16. doi: 10.1095/biolreprod.112.105619. [DOI] [PubMed] [Google Scholar]
- 79.Herrera E, Desoye G. Maternal and fetal lipid metabolism under normal and gestational diabetic conditions. Horm Mol Biol Clin Investig. 2016;26:109–127. doi: 10.1515/hmbci-2015-0025. [DOI] [PubMed] [Google Scholar]
- 80.Lindegaard ML, Wassif CA, Vaisman B, Amar M, Wasmuth EV, Shamburek R, et al. Characterization of placental cholesterol transport: ABCA1 is a potential target for in utero therapy of Smith–Lemli–Opitz syndrome. Hum Mol Genet. 2008;17:3806–3813. doi: 10.1093/hmg/ddn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindegaard MLS, Damm P, Mathiesen ER, Nielsen LB. Placental triglyceride accumulation in maternal type 1 diabetes is associated with increased lipase gene expression. J Lipid Res. 2006;47:2581–2588. doi: 10.1194/jlr.M600236-JLR200. [DOI] [PubMed] [Google Scholar]
- 82.Baardman ME, Kerstjens-Frederikse WS, Berger RMF, Bakker MK, Hofstra RMW, Plösch T. The role of maternal–fetal cholesterol transport in early fetal life: Current insights. Biol Reprod. 2013;88:24. doi: 10.1095/biolreprod.112.102442. [DOI] [PubMed] [Google Scholar]
- 83.Viau M, Lafond J, Vaillancourt C. Expression of placental serotonin transporter and 5-HT 2A receptor in normal and gestational diabetes mellitus pregnancies. Reprod Biomed Online. 2009;19:207–215. doi: 10.1016/s1472-6483(10)60074-0. [DOI] [PubMed] [Google Scholar]
- 84.Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, et al. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–350. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gabory A, Ferry L, Fajardy I, Jouneau L, Gothié J-D, Vigé A, et al. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One. 2012;7:e47986. doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 88.Ayala R, Shu T, Tsai L-H. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 89.da Silva JS, Dotti CG. Breaking the neuronal sphere: Regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 90.Jing Y-H, Song Y-F, Yao Y-M, Yin J, Wang D-G, Gao L-P. Retardation of fetal dendritic development induced by gestational hyperglycemia is associated with brain insulin/IGF-I signals. Int J Dev Neurosci. 2014;37:15–20. doi: 10.1016/j.ijdevneu.2014.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.