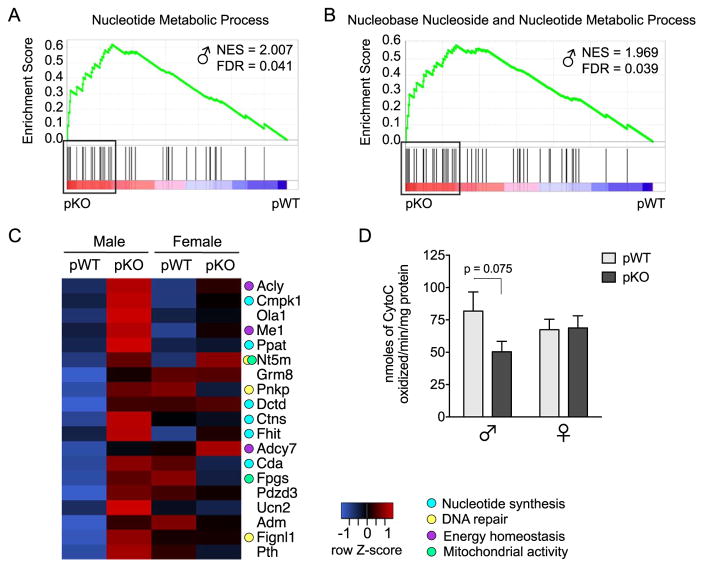
Figure 3.
Long-term prefrontal cortex (PFC) programming in male mice with placental insulin receptor (InsR) deficiency points to a disruption in cellular metabolic processes. (A, B) Gene set enrichment analysis (GSEA) of the adult PFC transcriptome in placental trophoblast-specific InsR deficiency (pKO) vs. control (pWT) male mice (n = 6/group) identified two significantly enriched gene sets in the pKO group, indicating programming of PFC nucleotide/nucleoside metabolic processes. Plots depict gene set enrichment score, normalized enrichment score (NES), distribution of set members across the GSEA-ranked gene list, and genes comprising the leading-edge subset (boxes). (C) Leading-edge analysis revealed an upregulation of genes involved in nucleotide synthesis, DNA repair, metabotropic glutamate receptor signaling, and mitochondrial activity in the PFC of adult pKO male mice. The heat map depicts expression of the leading-edge subset in the adult PFC, where blue indicates lower and red indicates higher average relative gene expression across groups. (D) Cytochrome C oxidase (CCO) activity in the PFC of experiment-naïve adults. As would be predicted from the GSEA results, a trending decrease in CCO activity was detected in pKO male mice (a priori t test, pWT vs. pKO male mice: t13 = 1.94, p = .075); however, these results did not reach statistical significance, interaction: F1,2 = 2.61, p = .12, genotype: F1,27 = 2.20, p = .15, sex: F1,27 = 0.039, p = .85. n = 7–8/group. FDR, false discovery rate.