Figure 4.
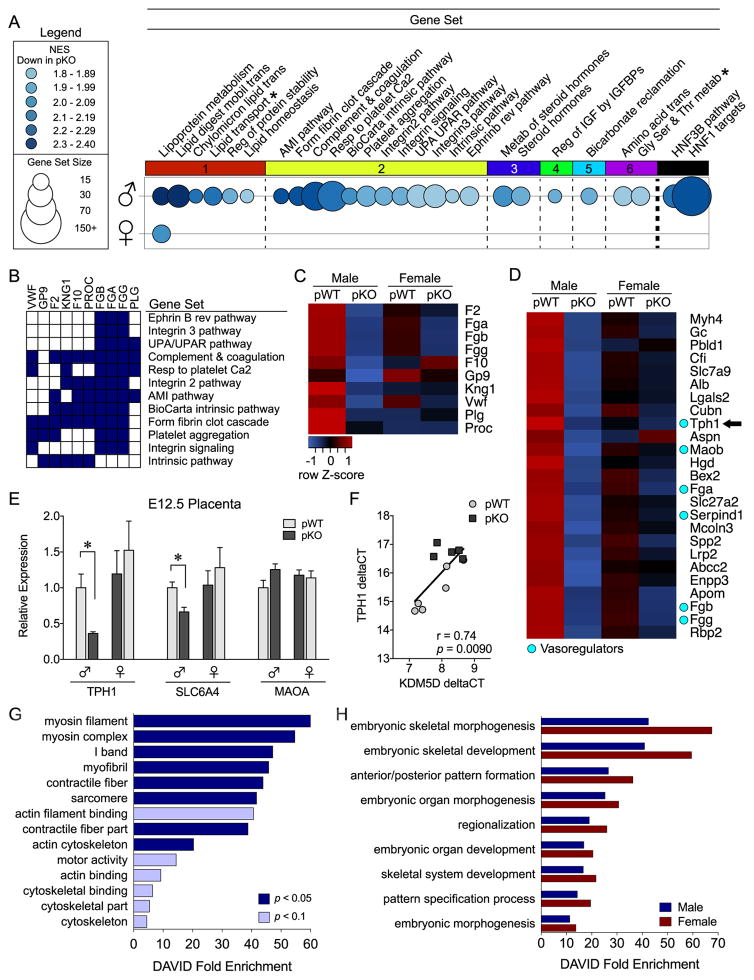
Insulin receptor (InsR) deficiency elicits sex-specific transcriptional responses in the placenta and fetal brain. (A–C) Gene set enrichment analysis of embryonic day (E) 17.5 placenta transcriptome identified key pathways dysregulated by InsR deletion (n = 6/group). (A) Bubble plot of gene sets (circles) significantly downregulated in male (top) and female (bottom) placental trophoblast-specific InsR deficiency (pKO) placentas relative to controls (pWT), where color intensity indicates the normalized enrichment score (NES) and diameter indicates gene set size. Differentially expressed gene sets (NES > 1.8, false discovery rate [FDR] < 0.05) were clustered by gene member overlap and ordered by NES. Whereas InsR deletion suppressed lipoprotein metabolism genes in both sexes, 25 additional gene sets were downregulated only in male pKO placentas. Clustering illustrated marked suppression of genes mediating (1) lipid homeostasis, (2) vascular function, (3) steroid hormone metabolism, (4) insulin-like growth factor (IGF) activity, (5) mitochondrial function, and (6) amino acid transport in pKO male mice. Gene set enrichment analysis (GSEA) also identified hepatocyte nuclear factor 1 (HNF1) as a candidate regulator in male mice, with the majority of gene sets containing HNF1 targets (* = not HNF1 target). Differentially expressed gene sets are detailed in Supplemental Table S2. (B) Plot showing the overlap of the leading-edge genes from each differentially expressed gene set in the vascular cluster. Expression of a common subset of coagulation cascade genes was suppressed in pKO male placentas. (C) A heat map of genes comprising the coagulation subset, where blue indicates lower and red indicates higher average relative expression across groups. InsR deletion decreased expression of procoagulant genes in male mice. (D) A heat map of the top 20 GSEA-ranked genes correlated with InsR deletion in male E17.5 placentas. Cyan circles indicate vasoregulators, including the rate-limiting serotonin synthesizing enzyme tryptophan hydroxylase 1 (TPH1, arrow). (E) Consistent with E17.5, TPH1 messenger RNA was suppressed by InsR knockout by E12.5 in male placentas (a priori t test, pWT vs. pKO male mice: t9 = 3.0, p = .015) but not in female mice (t10 = 0.63, p = .54), although the interaction between genotype and sex did not reach statistical significance, F1,19 = 2.75, p = .11. The serotonin reuptake transporter (SLC6A4) was similarly decreased in male tissue (a priori t test, pWT vs. pKO male mice: t9 = 3.25, p = .0087) but not in female mice (t10 = 0.71, p = .49), whereas no genotype by sex interaction was detected, F1,20 = 2.61, p = .12. Conversely, placental expression of monoamine oxidase A (MAOA) in male mice was not significantly affected by InsR deficiency (a priori t test, pWT vs. pKO male mice: t9 = 1.87, p = .095). (F) Expression of the Y-linked histone demethylase KDM5D was positively correlated with TPH1 in male E12.5 placentas (R2 9 = .55, p = .009). (G–H) Because placenta-derived serotonin is critical for fetal neurodevelopment, we assessed the E12.5 brain transcriptome for sex-specific effects of pKO. Functional annotation clustering of differentially expressed genes suggested disruption of motility and cytoskeletal dynamics only in pKO male mice (G), whereas the remaining effects of pKO were detected in both male and female mice (H). AMI, acute myocardial infarction; DAVID, Database for Annotation, Visualization, and Integrated Discovery; HNF3B, hepatocyte nuclear factor 3 beta; UPA, urokinase-type plasminogen activator; UPAR, UPA receptor.