Fig. 3.
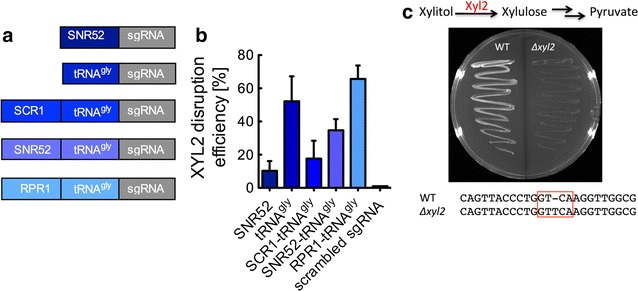
CRISPR–Cas9 genome editing in K. marxianus. a The design of native and hybrid RNA polymerase III promoters for sgRNA expression. The 20 bp sgRNA targeting sequence targets xylitol dehydrogenase (XYL2). b CRISPR–Cas9-induced disruption efficiencies of the XYL2 gene. Cultures were transformed with a single vector expressing codon-optimized Cas9 and the sgRNA. Cultures were grown on selective media for 2 days prior to screening on xylitol supplemented agar media. Data points represent the arithmetic mean of 30 colonies randomly selected from three different transformations. Error bars represent the standard deviation of the samples. c XYL2-disrupted strains were restreaked on xylitol-containing agar plates and screened for loss of growth. Gene disruption was confirmed by sequencing
